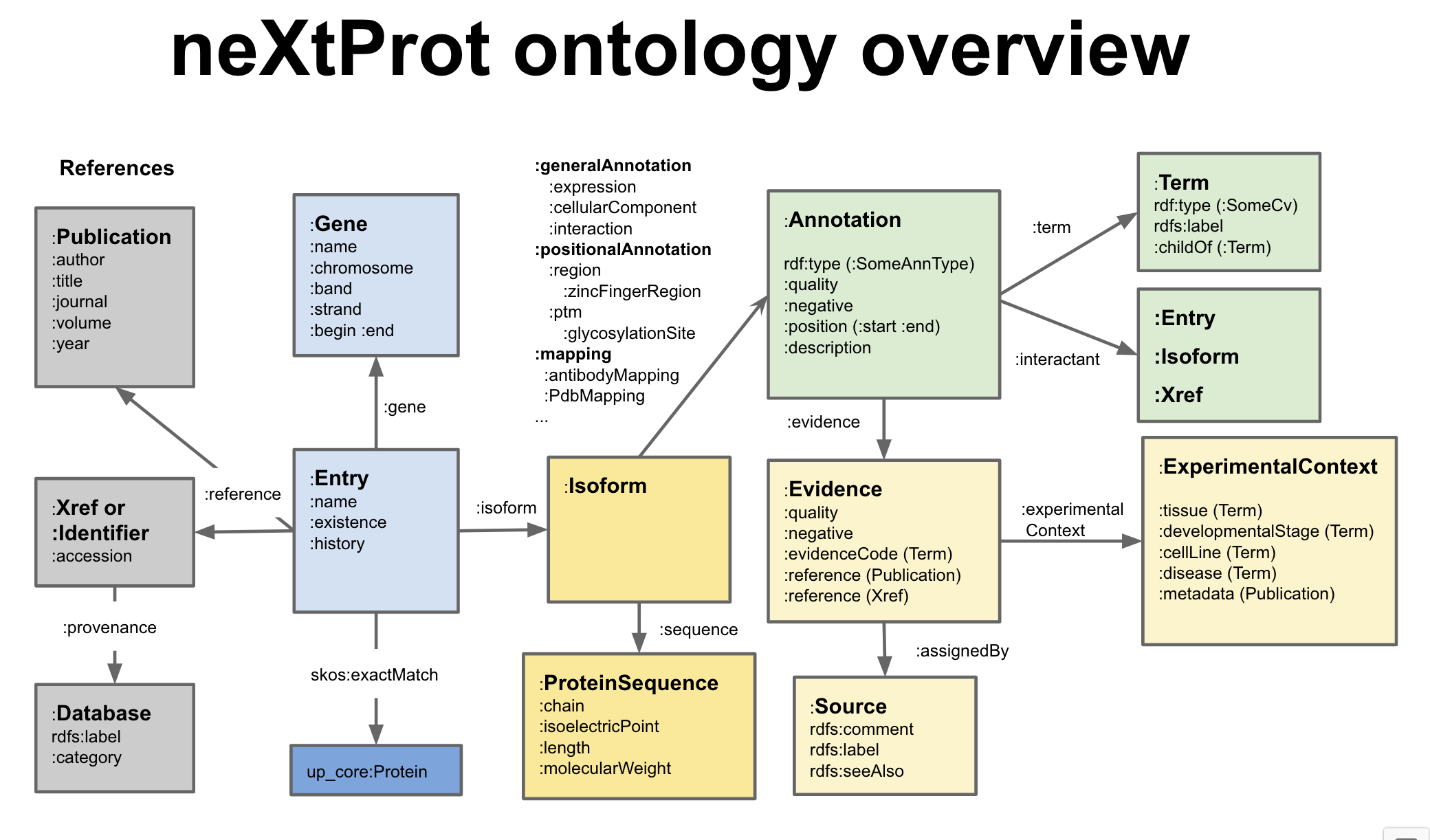
Pictures say 1,000 words
Figure 1: Ontology overview
http://nextprot.org/rdfThe neXtProt ontology is used to represent various evidence based statements (multiple annotation types) regarding human proteins

| URI | http://nextprot.org/rdf#AbsorptionMax |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Wavelength at which maximal light absorption by the photoreactive protein occurs. Other observed absorbance maxima (of lesser importance), lambda(max) for light emission, experimental or environmental remarks may also be included. |
| Super-classes |
Biophysicochemical propertyc |
| In range of |
absorption maxop |
| URI | http://nextprot.org/rdf#AbsorptionNote |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Note with additional information on light absorption by the photoreactive protein. |
| Super-classes |
Biophysicochemical propertyc |
| In range of |
absorption noteop |
| URI | http://nextprot.org/rdf#ActiveSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid(s) directly involved in the catalytic activity of the protein. |
| Super-classes |
Sitec |
| In range of |
active siteop |
| URI | http://nextprot.org/rdf#ActivityRegulation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Mechanisms which regulate the activity of the protein, including the components which regulate (by activation or inhibition) the reaction. |
| Super-classes |
Interactionc |
| In range of |
activity regulationop |
| URI | http://nextprot.org/rdf#Allergen |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Protein causing an allergic reaction. |
| Super-classes |
Medicalc |
| In range of |
allergenop |
| URI | http://nextprot.org/rdf#AllergenName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Allergen name. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#Annotation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
A statement about a protein. |
| Sub-classes |
Positional annotationc General Annotationc |
| In domain of |
evidenceop negative evidenceop |
| URI | http://nextprot.org/rdf#AntibodyMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Fragment of the protein or PrESTs (Protein Epitope Signature Tags) used to generate antibodies by the Human Protein Atlas project. |
| Super-classes |
Mappingc |
| In domain of |
antibody uniquenessdp antibody namedp |
| In range of |
antibody mappingop |
| URI | http://nextprot.org/rdf#BetaStrand |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Beta strand within the protein structure. |
| Super-classes |
Secondary structurec |
| In range of |
beta strandop |
| URI | http://nextprot.org/rdf#BgeeDevelopmentalStageCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the Bgee controlled vocabulary of human developmental stages. |
| Super-classes |
Termc |
| In range of |
developmental stageop |
| URI | http://nextprot.org/rdf#BinaryInteraction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Interaction involving two proteins. |
| Super-classes |
Interactionc |
| In domain of |
self-interactiondp |
| In range of |
binary interactionop |
| URI | http://nextprot.org/rdf#BindingSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Binding site for any chemical group (co-enzyme, prosthetic group, etc.). |
| Super-classes |
Sitec |
| In range of |
binding siteop |
| URI | http://nextprot.org/rdf#BiophysicochemicalProperty |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Biophysical and chemical properties of the protein, such as maximal absorption, kinetic parameters, pH dependence, redox potential and temperature dependence. |
| Super-classes |
General Annotationc |
| Sub-classes |
Redox potentialc Kinetic Vmaxc Temperature dependencec PH dependencec Kinetic KMc Absorption maxc Kinetic notec Absorption notec |
| In range of |
biophysicochemical propertyop |
| URI | http://nextprot.org/rdf#CDAntigenName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Antigen name according to the cluster of differentiation (CD) nomenclature. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#CalciumBindingRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Calcium binding region within the protein. |
| Super-classes |
Regionc |
| In range of |
calcium-binding regionop |
| URI | http://nextprot.org/rdf#CatalyticActivity |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Reaction(s) catalyzed by the protein. |
| Super-classes |
Functionc |
| In range of |
catalytic activityop |
| URI | http://nextprot.org/rdf#Caution |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Warning about possible errors and/or grounds for confusion. |
| Super-classes |
General Annotationc |
| In range of |
cautionop |
| URI | http://nextprot.org/rdf#CellularComponent |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Cellular component where the protein is localized. |
| Super-classes |
General Annotationc |
| Sub-classes |
Subcellular locationc Subcellular location informationc GO cellular componentc |
| In range of |
cellular componentop |
| URI | http://nextprot.org/rdf#CleavageSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Natural proteolytic cleavage site in the protein. |
| Super-classes |
Sitec |
| In range of |
cleavage siteop |
| URI | http://nextprot.org/rdf#CleavedRegionName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Cleaved region name. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#Cofactor |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Non-protein substance required for enzyme activity. |
| Super-classes |
Interactionc |
| In range of |
cofactorop |
| URI | http://nextprot.org/rdf#CofactorInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on protein cofactor(s). |
| Super-classes |
Interactionc |
| In range of |
cofactor informationop |
| URI | http://nextprot.org/rdf#CoiledCoilRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Coiled-coil region within the protein. |
| Super-classes |
Regionc |
| In range of |
coiled-coil regionop |
| URI | http://nextprot.org/rdf#CompositionallyBiasedRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region of compositional bias within the protein, in which a particular type of amino acid is over-represented. |
| Super-classes |
Regionc |
| In range of |
compositionally biased regionop |
| URI | http://nextprot.org/rdf#Consortium |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Name of a specific consortium, committee, or study group. |
| URI | http://nextprot.org/rdf#CrossLink |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid(s) participating in covalent linkage between proteins. |
| Super-classes |
PTMc |
| In range of |
cross-linkop |
| URI | http://nextprot.org/rdf#DevelopmentalStageInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information describing the expression of the gene product(s) at various stages of a cell, tissue or organism development. |
| Super-classes |
Expressionc |
| In range of |
developmental stage expression informationop |
| URI | http://nextprot.org/rdf#Disease |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Disease associated with variations in the gene encoding the protein. |
| Super-classes |
Medicalc |
| URI | http://nextprot.org/rdf#DisulfideBond |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Cysteine(s) participating in disulfide bonds. |
| Super-classes |
PTMc |
| In range of |
disulfide bondop |
| URI | http://nextprot.org/rdf#DnaBindingRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
DNA-binding region within the protein. |
| Super-classes |
Regionc |
| In range of |
dna-binding regionop |
| URI | http://nextprot.org/rdf#Domain |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region within the protein with a characteristic three-dimensional structure or fold. |
| Super-classes |
Regionc |
| In range of |
domainop |
| URI | http://nextprot.org/rdf#DomainInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information describing domain(s). |
| Super-classes |
General Annotationc |
| In range of |
domain informationop |
| URI | http://nextprot.org/rdf#ElectrophysiologicalParameter |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Biophysical properties of a ion channel. |
| Super-classes |
General Annotationc |
| URI | http://nextprot.org/rdf#Entry |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Sequence(s) of protein isoform(s) encoded by one or more human genes, and associated information. |
| In domain of |
existenceop swissprot pagedp family nameop isoform countdp isoformop historyop geneop |
| In range of |
best gene mappingop |
| URI | http://nextprot.org/rdf#EnzymeClassification |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Enzymatic activity of the protein annotated using the Enzyme Commission (EC) classification |
| Super-classes |
General Annotationc |
| In range of |
enzyme classificationop |
| URI | http://nextprot.org/rdf#EnzymeClassificationCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Number from the Enzyme Commission (EC) classification scheme for enzymes. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#EnzymeName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Name from the Enzyme Commission (EC) classification scheme for enzymes. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#Evidence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information supporting an annotation concerning the protein. |
| In domain of |
allele countdp experimental contextop homozygote allele countdp number of experimentsdp allele numberdp interaction detection methoddp negativedp expression scoredp from databaseop allele frequencydp go qualifierdp observed expressionop severitydp assigned byfp evidence codeop |
| In range of |
negative evidenceop evidenceop |
| URI | http://nextprot.org/rdf#EvidenceCodeOntologyCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the ECO controlled vocabulary of evidence and assertion methods. |
| Super-classes |
Termc |
| In range of |
evidence codeop |
| URI | http://nextprot.org/rdf#ExperimentalContext |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Experimental context in which the protein was studied. |
| In domain of |
metadatadp developmental stageop cell lineop tissueop |
| In range of |
experimental contextop |
| URI | http://nextprot.org/rdf#Expression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Expression of the gene product(s) in cells or in tissues. |
| Super-classes |
General Annotationc |
| Sub-classes |
Developmental stage expression informationc Expression informationc Expression profilec |
| In range of |
expressionop |
| URI | http://nextprot.org/rdf#ExpressionInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on the expression of the gene product(s) in cells or in tissues. |
| Super-classes |
Expressionc |
| In range of |
expression informationop |
| URI | http://nextprot.org/rdf#ExpressionProfile |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Expression of the gene product(s) in a given organ, tissue, cell type, fluid or gestational structure. |
| Super-classes |
Expressionc |
| In range of |
undetected expressionop detected expressionop medium expressionop expression profileop low expressionop high expressionop |
| URI | http://nextprot.org/rdf#FamilyName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on the family of proteins to which the protein belongs. |
| Super-classes |
Namec |
| In range of |
family nameop |
| URI | http://nextprot.org/rdf#Function |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Function(s) of the protein. |
| Super-classes |
General Annotationc |
| Sub-classes |
Pathwayc Catalytic activityc GO biological processc GO molecular functionc Function informationc Transport activityc |
| In range of |
functionop |
| URI | http://nextprot.org/rdf#FunctionInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on the function(s) of the protein. |
| Super-classes |
Functionc |
| In range of |
function informationop |
| URI | http://nextprot.org/rdf#FunctionalRegionName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Functional region name |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#Gene |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information about the gene(s) encoding the protein isoform sequence(s) described in the entry. |
| In domain of |
chromosomal banddp chromosomedp chromosome stranddp begindp best gene mappingop |
| In range of |
geneop |
| URI | http://nextprot.org/rdf#GeneName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Gene name. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#GeneralAnnotation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Annotation describing a biological aspect of the protein. |
| Super-classes |
Annotationc |
| Sub-classes |
Protein propertyc Medicalc PTM informationc Expressionc Inductionc Cautionc Cellular componentc Mammalian phenotypec Miscellaneousc Electrophysiological parameterc Variant informationc Domain informationc Phenotype variationc Interactionc Enzyme classificationc UniProtKB keyword annotationc Functionc Sequence cautionc Biophysicochemical propertyc |
| In range of |
general annotationop |
| URI | http://nextprot.org/rdf#GlycosylationSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid covalently attached to a glycan group (mono-, di-, or polysaccharide). |
| Super-classes |
PTMc |
| In range of |
glycosylation siteop |
| URI | http://nextprot.org/rdf#GoBiologicalProcess |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Involvement of the protein in a larger process, or ‘biological program’. |
| Super-classes |
Functionc |
| In range of |
go biological processop |
| URI | http://nextprot.org/rdf#GoBiologicalProcessCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the GO controlled vocabulary of biological processes. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#GoCellularComponent |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Location in the cell of the mature protein using the Gene Ontology cellular component vocabulary. |
| Super-classes |
Cellular componentc |
| In range of |
go cellular componentop |
| URI | http://nextprot.org/rdf#GoCellularComponentCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the GO controlled vocabulary of cellular components. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#GoMolecularFunction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Activity of the protein at the molecular level. |
| Super-classes |
Functionc |
| In range of |
go molecular functionop |
| URI | http://nextprot.org/rdf#GoMolecularFunctionCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the GO controlled vocabulary of molecular functions. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#GoQualifier |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
GO qualifier |
| In range of |
go qualifierdp |
| Members |
Located_in Contributes_to Part_of Is_active_in Enables Colocalizes_with |
| URI | http://nextprot.org/rdf#Helix |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Helical region within the protein structure. |
| Super-classes |
Secondary structurec |
| In range of |
helixop |
| URI | http://nextprot.org/rdf#History |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
History of the entry. |
| Sub-classes |
UniProt historyc neXtProt historyc |
| In domain of |
versiondp updateddp integrateddp |
| In range of |
historyop |
| URI | http://nextprot.org/rdf#Identifier |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Identifier |
| URI | http://nextprot.org/rdf#Induction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Effect of chemical compounds, environmental factors or transcriptional regulators on the expression of the protein. |
| Super-classes |
General Annotationc |
| In range of |
inductionop |
| URI | http://nextprot.org/rdf#InitiatorMethionine |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Initiator methionine cleaved during protein processing. |
| Super-classes |
Processing product.c |
| In range of |
initiator methionineop |
| URI | http://nextprot.org/rdf#InteractingRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Smallest region within the protein experimentally shown to participate in a given binary protein interaction. |
| Super-classes |
Regionc |
| In range of |
interacting regionop |
| URI | http://nextprot.org/rdf#Interaction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Interaction(s) of the protein with other proteins or molecules. |
| Super-classes |
General Annotationc |
| Sub-classes |
Small molecule interactionc Cofactorc Cofactor informationc Binary interactionc Activity regulationc Interaction informationc |
| In range of |
interactionop |
| URI | http://nextprot.org/rdf#InteractionInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on the protein quaternary structure and interaction(s) with other proteins or protein complexes. |
| Super-classes |
Interactionc |
| In range of |
interaction informationop |
| URI | http://nextprot.org/rdf#InteractionMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region within the protein shown experimentally to participate in a given binary protein interaction. |
| Super-classes |
Mappingc |
| In range of |
interaction mappingop |
| URI | http://nextprot.org/rdf#InternationalNonproprietaryName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
International non-proprietary name (INN) of a protein. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#IntramembraneRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region within the protein interacting with the membrane lipid bilayer. |
| Super-classes |
Topologyc |
| In range of |
intramembrane regionop |
| URI | http://nextprot.org/rdf#IsoformName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Protein isoform name. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#KineticKM |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Michaelis constant (KM). This is the substrate concentration at which the reaction rate is half of its maximal value. |
| Super-classes |
Biophysicochemical propertyc |
| In range of |
kinetic kmop |
| URI | http://nextprot.org/rdf#KineticNote |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Note with additional information on the kinetic data for the protein. |
| Super-classes |
Biophysicochemical propertyc |
| In range of |
kinetic noteop |
| URI | http://nextprot.org/rdf#KineticVmax |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Maximal velocity (Vmax). Vmax is the reaction rate attained when the enzyme sites are saturated with substrate. |
| Super-classes |
Biophysicochemical propertyc |
| In range of |
kinetic vmaxop |
| URI | http://nextprot.org/rdf#LargeScalePublication |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Publication relevant to 15 or more protein entries. |
| Super-classes |
Publicationc |
| URI | http://nextprot.org/rdf#LipidationSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid(s) covalently attached to a lipid group. |
| Super-classes |
PTMc |
| In range of |
lipid moiety-binding regionop |
| URI | http://nextprot.org/rdf#MammalianPhenotype |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Phenotype observed in a cell, tissue or organism. |
| Super-classes |
General Annotationc |
| URI | http://nextprot.org/rdf#MammalianPhenotypeCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the MGI controlled vocabulary of mammalian phenotypes. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#Mapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Mapping of sequences (PrEST, peptide, etc.) to the protein isoform sequence. |
| Super-classes |
Positional annotationc |
| Sub-classes |
PDB mappingc Interaction mappingc Peptide mappingc Antibody mappingc SRM peptide mappingc |
| In range of |
mappingop |
| URI | http://nextprot.org/rdf#MatureProtein |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Mature form of the protein. Processing (signal peptide and propeptide cleavage, transit peptide cleavage or initiator methionine cleavage) is required for maturation. |
| Super-classes |
Processing product.c |
| In range of |
mature proteinop |
| URI | http://nextprot.org/rdf#Medical |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Medically-relevant information on the protein. |
| Super-classes |
General Annotationc |
| Sub-classes |
Allergenc Diseasec Pharmaceuticalc |
| In range of |
medicalop |
| URI | http://nextprot.org/rdf#MeshAnatomyCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the NLM controlled vocabulary of medical subject headings (MeSH), A part (anatomy). |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#MeshCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the NLM controlled vocabulary of medical subject headings (MeSH). |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#MetalBindingSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid directly involved in binding a metal ion. |
| Super-classes |
Sitec |
| In range of |
metal binding siteop |
| URI | http://nextprot.org/rdf#Miscellaneous |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on the protein that doesn't fall into the scope of any other section. |
| Super-classes |
General Annotationc |
| In range of |
miscellaneousop |
| URI | http://nextprot.org/rdf#MiscellaneousRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region of interest within the protein. |
| Super-classes |
Regionc |
| In range of |
miscellaneous regionop |
| URI | http://nextprot.org/rdf#MiscellaneousSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid of interest within the protein. |
| Super-classes |
Sitec |
| In range of |
miscellaneous siteop |
| URI | http://nextprot.org/rdf#MitochondrialTransitPeptide |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Transit peptide responsible for the transport of the protein to mitochondria. |
| Super-classes |
Processing product.c |
| In range of |
mitochondrial transit peptideop |
| URI | http://nextprot.org/rdf#ModifiedResidue |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid modified post-translationally (excluding lipids, glycans and protein cross-links). |
| Super-classes |
PTMc |
| In range of |
modified residueop |
| URI | http://nextprot.org/rdf#Mutagenesis |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid which has been experimentally altered by mutagenesis. |
| Super-classes |
Positional annotationc |
| In range of |
mutagenesisop |
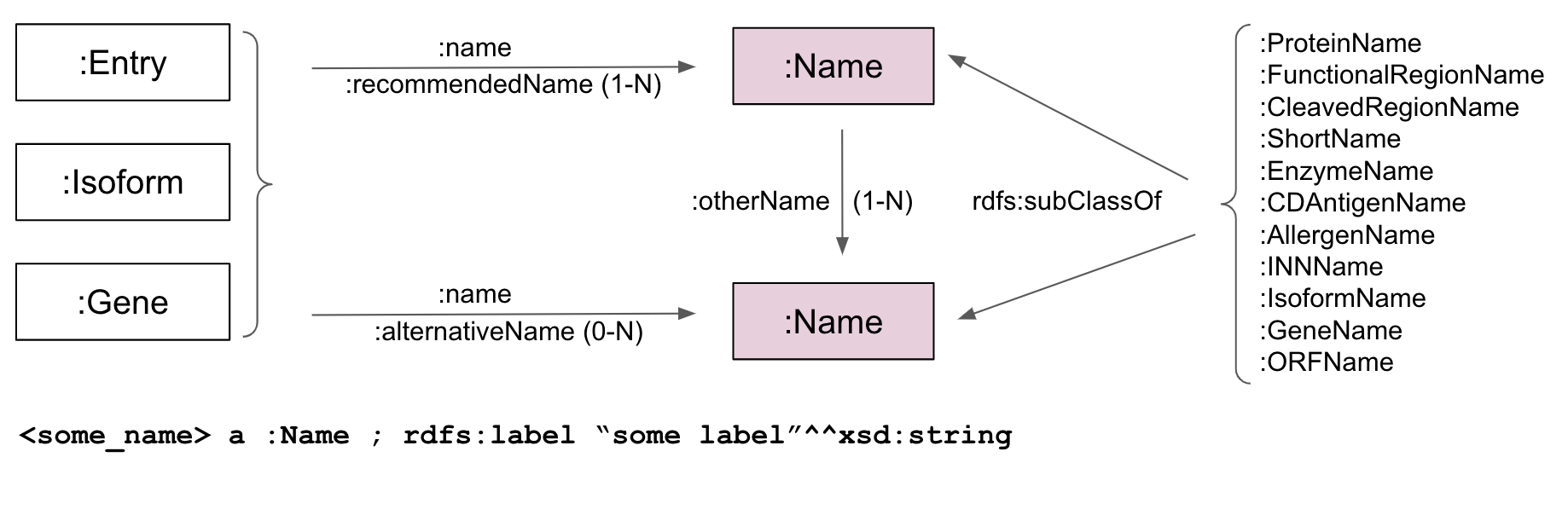
| URI | http://nextprot.org/rdf#Name |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Name. |
| Sub-classes |
Gene namec Cleaved region namec Family namec ORF namec Short namec Enzyme classification namec CD Antigen namec International Non proprietary Namec Functional region namec Protein namec Protein isoform namec Allergen namec |
| In domain of |
other nameop |
| In range of |
recommended nameop other nameop alternative nameop |
| URI | http://nextprot.org/rdf#NciMetathesaurusCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the NCI Metathesaurus controlled vocabulary. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NciThesaurusCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the NCI Thesaurus controlled vocabulary. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NextprotAnatomyCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the neXtProt controlled vocabulary of human anatomy and cell types. |
| Super-classes |
Termc |
| In range of |
tissueop |
| URI | http://nextprot.org/rdf#NextprotCellosaurusCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the Cellosaurus controlled vocabulary of cell lines. |
| Super-classes |
Termc |
| In range of |
cell lineop |
| URI | http://nextprot.org/rdf#NextprotDomainCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the neXtProt controlled vocabulary of domains, repeat and zinc fingers in human proteins (derived from UniProt). |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NextprotFamilyCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the neXtProt list of human protein families (derived from UniProt). |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NextprotHistory |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
neXtProt entry history |
| Super-classes |
Protein record historyc |
| URI | http://nextprot.org/rdf#NextprotIcepoCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the neXtProt controlled vocabulary of ion channel electrophysiology (ICEPO). |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NextprotMetalCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the neXtProt controlled vocabulary of metal bound to human proteins (derived from UniProt). |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NextprotModificationEffectCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the neXtProt controlled vocabulary describing changes resulting from a modification on a protein. Possible modifications include variations and post-translational modifications, while changes can be for example a sub-cellular localization, a phenotype, or a protein property. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NextprotProteinPropertyCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the neXtProt controlled vocabulary of protein properties. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NextprotTopologyCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the neXtProt controlled vocabulary describing the topology of transmembrane protein regions that span membrane compartments (derived from UniProt). |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NonConsecutiveResidue |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Two amino acids which are not consecutive and with a undetermined number of unsequenced residues between them. |
| Super-classes |
Positional annotationc |
| In range of |
non-consecutive residueop |
| URI | http://nextprot.org/rdf#NonStandardAminoAcidCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the controlled vocabulary of non-standard amino acids in protein sequences. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#NonTerminalResidue |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid which is not the terminal residue of the complete protein sequence. The sequence is incomplete. |
| Super-classes |
Positional annotationc |
| In range of |
non-terminal residueop |
| URI | http://nextprot.org/rdf#NucleotidePhosphateBindingRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region binding nucleotide phosphates. |
| Super-classes |
Regionc |
| In range of |
nucleotide phosphate-binding regionop |
| URI | http://nextprot.org/rdf#ORFName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Open reading frame (ORF) name of the gene encoding the protein isoform(s). |
| Super-classes |
Namec |
| In range of |
orf nameop |
| URI | http://nextprot.org/rdf#ObservedExpression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Observed expression (:Positive :High :Medium :Low :Negative) of the gene product(s). |
| In range of |
observed expressionop |
| Members |
Positive High Negative Low Medium |
| URI | http://nextprot.org/rdf#OmimCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the OMIM controlled vocabulary of human genes and genetic phenotypes. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#OrganelleCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the controlled vocabulary of organelles to which transit peptides can transport proteins. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#Pathway |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Series of actions involving the protein and other molecules in a cell that leads to a certain product or a change in the cell. |
| Super-classes |
Functionc |
| In range of |
pathwayop |
| URI | http://nextprot.org/rdf#PdbMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region of the protein for which there is a 3D structure in PDB. |
| Super-classes |
Mappingc |
| In domain of |
methoddp |
| In range of |
pdb mappingop |
| URI | http://nextprot.org/rdf#PeptideMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Peptide detected in a biological sample using mass spectrometry (MS). |
| Super-classes |
Mappingc |
| In domain of |
peptide sourcedp |
| In range of |
peptide mappingop |
| URI | http://nextprot.org/rdf#PeroxisomeTransitPeptide |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Transit peptide responsible for the transport of the protein to peroxisomes. |
| Super-classes |
Processing product.c |
| In range of |
peroxisome transit peptideop |
| URI | http://nextprot.org/rdf#Person |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Person. |
| In domain of |
suffixdp |
| In range of |
editorop |
| URI | http://nextprot.org/rdf#PhDependence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Optimum pH for protein activity, the variation of activity with pH variation and the pH stability of the protein. |
| Super-classes |
Biophysicochemical propertyc |
| In range of |
pH dependenceop |
| URI | http://nextprot.org/rdf#Pharmaceutical |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Use of the protein as a pharmaceutical drug. |
| Super-classes |
Medicalc |
| In range of |
pharmaceuticalop |
| URI | http://nextprot.org/rdf#PhenotypicVariation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Variation in observed molecular or organism phenotype due to modifications to the protein. |
| Super-classes |
General Annotationc |
| In range of |
phenotype variationop |
| URI | http://nextprot.org/rdf#PositionalAnnotation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Annotation relevant to one or more amino acid(s) of the protein. |
| Super-classes |
Annotationc |
| Sub-classes |
Mutagenesisc Processing product.c Regionc Mappingc Non-terminal residuec Sequence conflictc Variantc Disease-related variantc Non-consecutive residuec PTMc Secondary structurec Topologyc Sitec |
| In domain of |
startdp |
| In range of |
positional annotationop |
| URI | http://nextprot.org/rdf#ProcessingProduct |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Part of the protein resulting from natural processing events. |
| Super-classes |
Positional annotationc |
| Sub-classes |
Signal peptidec Mitochondrial transit peptidec Initiator methioninec Mature proteinc Maturation peptidec Peroxisome transit peptidec |
| In range of |
processing productop |
| URI | http://nextprot.org/rdf#Propeptide |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Part of the protein that is cleaved during maturation or activation. A propeptide generally has no independent biological function. |
| Super-classes |
Processing product.c |
| In range of |
maturation peptideop |
| URI | http://nextprot.org/rdf#ProteinExistence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Type of evidence that supports the existence of the protein. |
| In domain of |
protein existence leveldp |
| In range of |
existenceop |
| Members |
Predicted Evidence_at_transcript_level Evidence_at_protein_level Inferred_from_homology Uncertain |
| URI | http://nextprot.org/rdf#ProteinName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Protein name. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#ProteinProperty |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
A property of the protein, such as stability. |
| Super-classes |
General Annotationc |
| URI | http://nextprot.org/rdf#ProteinSequence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Amino acid sequence of the protein isoform and its associated properties (length, molecular weight and isoelectric point) |
| In domain of |
molecular weightdp isoelectric pointdp chaindp |
| In range of |
protein sequenceop |
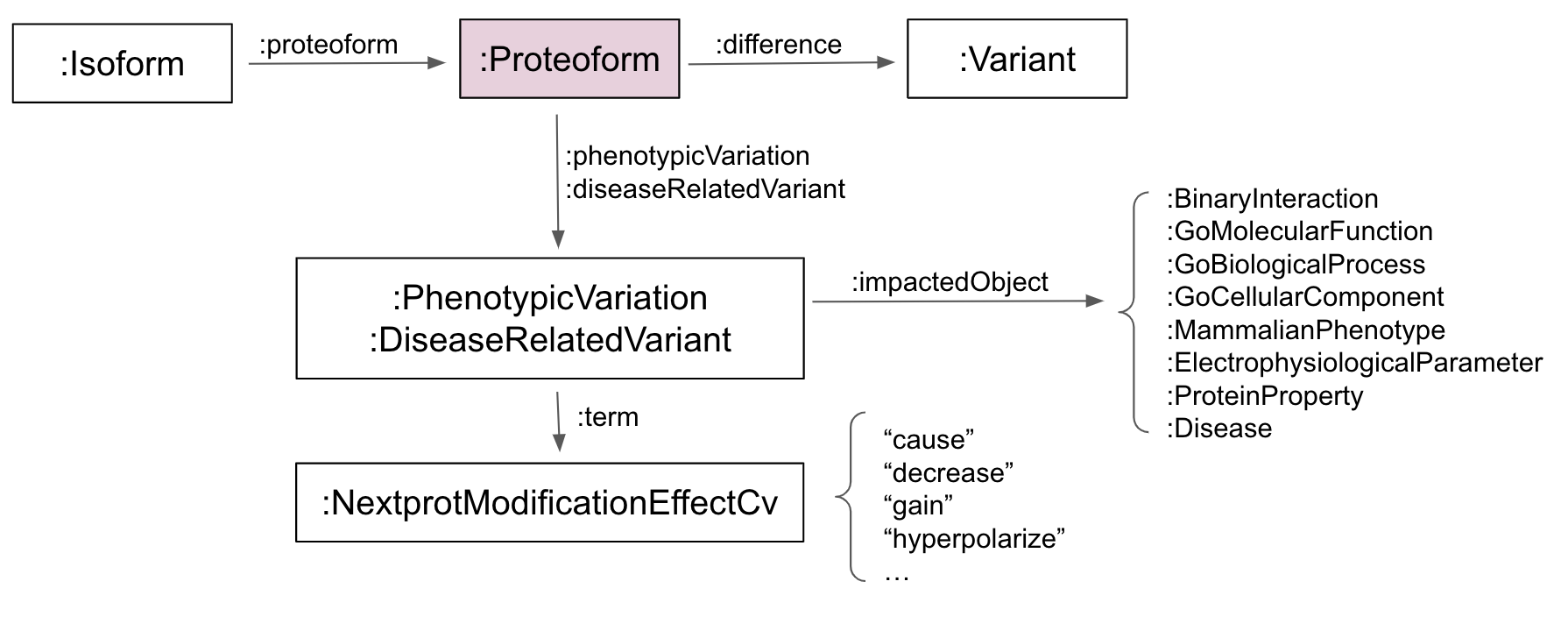
| URI | http://nextprot.org/rdf#Proteoform |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Protein isoform which include modifications, such as variant(s) and PTM(s). |
| Super-classes |
Isoformc |
| In domain of |
phenotype variationop differenceop disease-related variantop |
| In range of |
proteoformop |
| URI | http://nextprot.org/rdf#PsiMiCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the PSI-MI controlled vocabulary of molecular interactions. |
| Super-classes |
Termc |
| In range of |
interaction detection methoddp |
| URI | http://nextprot.org/rdf#Ptm |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Post-translational modification(s) of the protein. |
| Super-classes |
Positional annotationc |
| Sub-classes |
Selenocysteinec Cross-linkc Disulfide bondc Lipid moiety-binding regionc Modified residuec Glycosylation sitec |
| In range of |
ptmop |
| URI | http://nextprot.org/rdf#PtmInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on the post-translational modifications (PTMs) of the protein. |
| Super-classes |
General Annotationc |
| In range of |
ptm informationop |
| URI | http://nextprot.org/rdf#Publication |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Publication in the scientific literature. |
| Sub-classes |
Large scale publicationc |
| In domain of |
editorop fromdp issuedp last pagedp publication typedp volumedp citydp journaldp titledp publisherdp yeardp first pagedp authorop |
| In range of |
metadatadp |
| URI | http://nextprot.org/rdf#QualityQualifier |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Quality qualifier describing the reliability of an annotation or evidence. |
| In range of |
qualityop |
| Members |
BRONZE GOLD SILVER |
| URI | http://nextprot.org/rdf#RedoxPotential |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Value of the standard (midpoint) oxido-reduction potential(s) for the electron transport protein. |
| Super-classes |
Biophysicochemical propertyc |
| In range of |
redox potentialop |
| URI | http://nextprot.org/rdf#Region |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region of the protein isoform. |
| Super-classes |
Positional annotationc |
| Sub-classes |
Miscellaneous regionc Coiled-coil regionc Calcium-binding regionc DNA-binding regionc Repeatc Domainc Zinc finger regionc Nucleotide phosphate-binding regionc Interacting regionc Compositionally biased regionc Short sequence motifc |
| In range of |
regionop |
| URI | http://nextprot.org/rdf#Repeat |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Repeated sequence motifs or domains within the protein isoform. |
| Super-classes |
Regionc |
| In range of |
repeatop |
| URI | http://nextprot.org/rdf#SecondaryStructure |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Secondary structure (helix, turn, beta strand) within the protein. |
| Super-classes |
Positional annotationc |
| Sub-classes |
Beta strandc Turnc Helixc |
| In range of |
secondary structureop |
| URI | http://nextprot.org/rdf#Selenocysteine |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Selenocysteine present in the sequence of the protein. |
| Super-classes |
PTMc |
| In range of |
selenocysteineop |
| URI | http://nextprot.org/rdf#SequenceCaution |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on difference(s) between the protein isoform sequence(s) shown and other available protein sequences derived from the same gene. |
| Super-classes |
General Annotationc |
| In range of |
sequence cautionop |
| URI | http://nextprot.org/rdf#SequenceConflict |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Single amino acid difference between the sequence shown and another submitted sequence that cannot be explained by a known polymorphism. |
| Super-classes |
Positional annotationc |
| In range of |
sequence conflictop |
| URI | http://nextprot.org/rdf#Severity |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Level of severity of a phenotype variation |
| In range of |
severitydp |
| Members |
Severe Mild Moderate |
| URI | http://nextprot.org/rdf#ShortName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Shorter version of a name. |
| Super-classes |
Namec |
| URI | http://nextprot.org/rdf#ShortSequenceMotif |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Short sequence motif (up to 20 amino acids) of biological interest in the protein. |
| Super-classes |
Regionc |
| In range of |
short sequence motifop |
| URI | http://nextprot.org/rdf#SignalPeptide |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
N-terminal sequence targeting the protein to the secretory pathway. |
| Super-classes |
Processing product.c |
| In range of |
signal peptideop |
| URI | http://nextprot.org/rdf#Site |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Specific amino acid confering a property to the protein. |
| Super-classes |
Positional annotationc |
| Sub-classes |
Binding sitec Cleavage sitec Miscellaneous sitec Metal binding sitec Active sitec |
| In range of |
siteop |
| URI | http://nextprot.org/rdf#SmallMoleculeInteraction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Interaction of the protein with chemical molecule(s). |
| Super-classes |
Interactionc |
| In range of |
small molecule interactionop |
| URI | http://nextprot.org/rdf#SrmPeptideMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Peptide that can be used to quantify the protein by selected reaction monitoring (SRM) . |
| Super-classes |
Mappingc |
| In range of |
srm peptide mappingop |
| URI | http://nextprot.org/rdf#SubcellularLocation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Location in the cell of the mature protein using the UniProt vocabulary. |
| Super-classes |
Cellular componentc |
| In range of |
subcellular locationop |
| URI | http://nextprot.org/rdf#SubcellularLocationNote |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Note with additional information on the location in the cell of the mature protein. |
| Super-classes |
Cellular componentc |
| In range of |
subcellular location informationop |
| URI | http://nextprot.org/rdf#TemperatureDependence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Optimal temperature for enzyme activity and/or the variation of enzyme activity with temperature variation. The thermostability of the enzyme is mentioned, when known. |
| Super-classes |
Biophysicochemical propertyc |
| In range of |
temperature dependenceop |
| URI | http://nextprot.org/rdf#TopologicalDomain |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Location of non-membrane regions within a membrane-spanning protein. |
| Super-classes |
Topologyc |
| In range of |
topological domainop |
| URI | http://nextprot.org/rdf#Topology |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Description of how a transmembrane protein spans the membrane |
| Super-classes |
Positional annotationc |
| Sub-classes |
Topological domainc Intramembrane regionc Transmembrane regionc |
| In range of |
topologyop |
| URI | http://nextprot.org/rdf#TransmembraneRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Region of the protein which spans the membrane. |
| Super-classes |
Topologyc |
| In range of |
transmembrane regionop |
| URI | http://nextprot.org/rdf#TransportActivity |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Transport of substances by the protein isoform, usually across a membrane. |
| Super-classes |
Functionc |
| In range of |
transport activityop |
| URI | http://nextprot.org/rdf#Turn |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Hydrogen-bonded turn within the protein sequence. |
| Super-classes |
Secondary structurec |
| In range of |
turnop |
| URI | http://nextprot.org/rdf#UnipathwayCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the UniProt controlled vocabulary of metabolic pathways. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#UniprotDiseaseCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the UniProt controlled vocabulary of human diseases associated with genetic variations. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#UniprotHistory |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Uniprot entry history |
| Super-classes |
Protein record historyc |
| In domain of |
last sequence updatedp sequence versiondp |
| URI | http://nextprot.org/rdf#UniprotKeyword |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
UniProt keyword summarising the content of an entry. |
| Super-classes |
General Annotationc |
| In range of |
uniprot keywordop |
| URI | http://nextprot.org/rdf#UniprotKeywordCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
UniProt keyword. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#UniprotPtmCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the UniProt controlled vocabulary of post-translational modifications. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#UniprotSubcellularLocationCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the UniProt controlled vocabulary of subcellular locations. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#UniprotSubcellularOrientationCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the UniProt controlled vocabulary of subcellular locations describing a subcellular orientation. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#UniprotSubcellularTopologyCv |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Term from the UniProt controlled vocabulary of subcellular locations describing a subcellular topology. |
| Super-classes |
Termc |
| URI | http://nextprot.org/rdf#Variant |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Sequence variation observed in an individual which may or may not be linked to a disease. |
| Super-classes |
Positional annotationc |
| In range of |
variantop |
| URI | http://nextprot.org/rdf#VariantInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Information on polymorphic variant(s) of the protein isoform sequence. |
| Super-classes |
General Annotationc |
| In range of |
variant informationop |
| URI | http://nextprot.org/rdf#Version |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Version of the neXtProt API used to generate current data. |
| URI | http://nextprot.org/rdf#Xref |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Link to an external resource providing additional information. |
| In range of |
fromdp |
| URI | http://nextprot.org/rdf#ZincFingerRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Zinc finger region within the protein. |
| Super-classes |
Regionc |
| In range of |
zinc finger regionop |
| URI | http://nextprot.org/rdf#absorptionMax |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an absorption max annotation |
| Super-properties | biophysicochemical propertyop |
| Domain(s) | Isoformc |
| Range(s) | AbsorptionMaxc |
| URI | http://nextprot.org/rdf#absorptionNote |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an absorption note annotation |
| Super-properties | biophysicochemical propertyop |
| Domain(s) | Isoformc |
| Range(s) | AbsorptionNotec |
| URI | http://nextprot.org/rdf#activeSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an active site annotation |
| Super-properties | siteop |
| Domain(s) | Isoformc |
| Range(s) | ActiveSitec |
| URI | http://nextprot.org/rdf#activityRegulation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an activity regulation annotation |
| Super-properties | interactionop |
| Domain(s) | Isoformc |
| Range(s) | ActivityRegulationc |
| URI | http://nextprot.org/rdf#allergen |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an allergen annotation |
| Super-properties | medicalop |
| Domain(s) | Isoformc |
| Range(s) | Allergenc |
| URI | http://nextprot.org/rdf#alternativeName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein, gene or isoform to an alternative name |
| Super-properties | nameop |
| Domain(s) | (Proteinc or Isoformc or Genec) |
| Range(s) | Namec |
| URI | http://nextprot.org/rdf#antibodyMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an antibody mapping annotation |
| Super-properties | mappingop |
| Domain(s) | Isoformc |
| Range(s) | AntibodyMappingc |
| URI | http://nextprot.org/rdf#author |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to an author |
| Domain(s) | Publicationc |
| Range(s) | http://nextprot.org/rdf#Person http://nextprot.org/rdf#Consortium |
| URI | http://nextprot.org/rdf#bestGeneMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a gene to it being chosen as the representative gene for a multigenic entry |
| Domain(s) | Genec |
| Range(s) | Entryc |
| URI | http://nextprot.org/rdf#betaStrand |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a beta strand annotation |
| Super-properties | secondary structureop |
| Domain(s) | Isoformc |
| Range(s) | BetaStrandc |
| URI | http://nextprot.org/rdf#binaryInteraction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a binary interaction annotation |
| Super-properties | interactionop |
| Domain(s) | Isoformc |
| Range(s) | BinaryInteractionc |
| URI | http://nextprot.org/rdf#bindingSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a binding site annotation |
| Super-properties | siteop |
| Domain(s) | Isoformc |
| Range(s) | BindingSitec |
| URI | http://nextprot.org/rdf#biophysicochemicalProperty |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a biophysicochemical property annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | BiophysicochemicalPropertyc |
| URI | http://nextprot.org/rdf#calciumBindingRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a calcium-binding region annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | CalciumBindingRegionc |
| URI | http://nextprot.org/rdf#catalyticActivity |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a catalytic activity annotation |
| Super-properties | functionop |
| Domain(s) | Isoformc |
| Range(s) | CatalyticActivityc |
| URI | http://nextprot.org/rdf#caution |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a caution annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | Cautionc |
| URI | http://nextprot.org/rdf#cellLine |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an experimental context to a cell line |
| Domain(s) | Experimental contextc |
| Range(s) | NextprotCellosaurusCvc |
| URI | http://nextprot.org/rdf#cellularComponent |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a cellular component annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | CellularComponentc |
| URI | http://nextprot.org/rdf#cleavageSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a cleavage site annotation |
| Super-properties | siteop |
| Domain(s) | Isoformc |
| Range(s) | CleavageSitec |
| URI | http://nextprot.org/rdf#cofactor |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a cofactor annotation |
| Super-properties | interactionop |
| Domain(s) | Isoformc |
| Range(s) | Cofactorc |
| URI | http://nextprot.org/rdf#cofactorInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a cofactor information annotation |
| Super-properties | interactionop |
| Domain(s) | Isoformc |
| Range(s) | CofactorInfoc |
| URI | http://nextprot.org/rdf#coiledCoilRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a coiled-coil region annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | CoiledCoilRegionc |
| URI | http://nextprot.org/rdf#compositionallyBiasedRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a compositionally biased region annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | CompositionallyBiasedRegionc |
| URI | http://nextprot.org/rdf#crossLink |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a cross-link annotation |
| Super-properties | ptmop |
| Domain(s) | Isoformc |
| Range(s) | CrossLinkc |
| URI | http://nextprot.org/rdf#detectedExpression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to at least one detectable level of expression |
| Super-properties | expressionop |
| Domain(s) | Isoformc |
| Range(s) | ExpressionProfilec |
| URI | http://nextprot.org/rdf#developmentalStage |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an experimental context to a developmental stage |
| Domain(s) | Experimental contextc |
| Range(s) | BgeeDevelopmentalStageCvc |
| URI | http://nextprot.org/rdf#developmentalStageInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a developmental stage information annotation |
| Super-properties | expressionop |
| Domain(s) | Isoformc |
| Range(s) | DevelopmentalStageInfoc |
| URI | http://nextprot.org/rdf#difference |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a proteoform to a variant or mutagenesis annotation |
| Domain(s) | Proteoformc |
| Range(s) | http://nextprot.org/rdf#Mutagenesis http://nextprot.org/rdf#Variant |
| URI | http://nextprot.org/rdf#disease |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a disease annotation or an experimental context to a disease term |
| Super-properties | medicalop |
| Domain(s) | (Experimental contextc or Isoformc) |
| Range(s) | http://nextprot.org/rdf#NciMetathesaurusCv http://nextprot.org/rdf#NciThesaurusCv http://nextprot.org/rdf#Disease |
| URI | http://nextprot.org/rdf#disulfideBond |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a disulfide bond annotation |
| Super-properties | ptmop |
| Domain(s) | Isoformc |
| Range(s) | DisulfideBondc |
| URI | http://nextprot.org/rdf#dnaBindingRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a DNA-binding region annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | DnaBindingRegionc |
| URI | http://nextprot.org/rdf#domain |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a domain annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | Domainc |
| URI | http://nextprot.org/rdf#domainInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a domain information annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | DomainInfoc |
| URI | http://nextprot.org/rdf#editor |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to an editor |
| Domain(s) | Publicationc |
| Range(s) | Personc |
| URI | http://nextprot.org/rdf#enzymeClassification |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an enzyme classification annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | EnzymeClassificationc |
| URI | http://nextprot.org/rdf#evidence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an annotation to an evidence |
| Domain(s) | Annotationc |
| Range(s) | Evidencec |
| URI | http://nextprot.org/rdf#evidenceCode |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence to an evidence code |
| Domain(s) | Evidencec |
| Range(s) | EvidenceCodeOntologyCvc |
| URI | http://nextprot.org/rdf#existence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein entry to a protein existence level |
| Domain(s) | Proteinc |
| Range(s) | ProteinExistencec |
| URI | http://nextprot.org/rdf#experimentalContext |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence to an experimental context |
| Domain(s) | Evidencec |
| Range(s) | ExperimentalContextc |
| URI | http://nextprot.org/rdf#expression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an expression annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | Expressionc |
| URI | http://nextprot.org/rdf#expressionInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an expression information annotation |
| Super-properties | expressionop |
| Domain(s) | Isoformc |
| Range(s) | ExpressionInfoc |
| URI | http://nextprot.org/rdf#expressionProfile |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an expression profile annotation |
| Super-properties | expressionop |
| Domain(s) | Isoformc |
| Range(s) | ExpressionProfilec |
| URI | http://nextprot.org/rdf#familyName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein to a family name annotation |
| Super-properties | nameop |
| Domain(s) | Proteinc |
| Range(s) | FamilyNamec |
| URI | http://nextprot.org/rdf#fromDatabase |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence to the database of the cross reference that the evidence is based on |
| Domain(s) | Evidencec |
| Range(s) | Databasec |
| URI | http://nextprot.org/rdf#function |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a function annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | Functionc |
| URI | http://nextprot.org/rdf#functionInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a function information annotation |
| Super-properties | functionop |
| Domain(s) | Isoformc |
| Range(s) | FunctionInfoc |
| URI | http://nextprot.org/rdf#gene |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein to its encoding gene |
| Domain(s) | Proteinc |
| Range(s) | Genec |
| URI | http://nextprot.org/rdf#generalAnnotation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a general annotation (function, expression, etc.) |
| Domain(s) | Isoformc |
| Range(s) | GeneralAnnotationc |
| URI | http://nextprot.org/rdf#glycosylationSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a glycosylation site annotation |
| Super-properties | ptmop |
| Domain(s) | Isoformc |
| Range(s) | GlycosylationSitec |
| URI | http://nextprot.org/rdf#goBiologicalProcess |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a GO biological process annotation |
| Super-properties | functionop |
| Domain(s) | Isoformc |
| Range(s) | GoBiologicalProcessc |
| URI | http://nextprot.org/rdf#goCellularComponent |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a GO cellular component annotation |
| Super-properties | cellular componentop |
| Domain(s) | Isoformc |
| Range(s) | GoCellularComponentc |
| URI | http://nextprot.org/rdf#goMolecularFunction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a GO molecular function annotation |
| Super-properties | functionop |
| Domain(s) | Isoformc |
| Range(s) | GoMolecularFunctionc |
| URI | http://nextprot.org/rdf#helix |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a helix annotation |
| Super-properties | secondary structureop |
| Domain(s) | Isoformc |
| Range(s) | Helixc |
| URI | http://nextprot.org/rdf#highExpression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a high level of expression |
| Super-properties | detected expressionop |
| Domain(s) | Isoformc |
| Range(s) | ExpressionProfilec |
| URI | http://nextprot.org/rdf#history |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein entry to its history in UniProt or neXtProt |
| Domain(s) | Proteinc |
| Range(s) | Historyc |
| URI | http://nextprot.org/rdf#impactedObject |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a phenotype variation annotation to an abnormal mammalian phenotype, electrophysiological parameter, binary interaction, protein property or GO annotation, or a disease-related variant to a disease |
| Domain(s) | (Disease-related variantc or Phenotype variationc) |
| Range(s) | http://nextprot.org/rdf#MammalianPhenotype http://nextprot.org/rdf#GoMolecularFunction http://nextprot.org/rdf#ElectrophysiologicalParameter http://nextprot.org/rdf#GoBiologicalProcess http://nextprot.org/rdf#GoCellularComponent http://nextprot.org/rdf#ProteinProperty http://nextprot.org/rdf#BinaryInteraction http://nextprot.org/rdf#Disease |
| URI | http://nextprot.org/rdf#induction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an induction annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | Inductionc |
| URI | http://nextprot.org/rdf#initiatorMethionine |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an initiator methionine annotation |
| Super-properties | processing productop |
| Domain(s) | Isoformc |
| Range(s) | InitiatorMethioninec |
| URI | http://nextprot.org/rdf#interactant |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an interactant (an entry, isoform or cross-reference) |
| Domain(s) | (Binary interactionc or Small molecule interactionc or Cofactorc or Interaction mappingc or Binding sitec) |
| Range(s) | http://nextprot.org/rdf#Xref http://nextprot.org/rdf#Entry http://nextprot.org/rdf#Isoform |
| URI | http://nextprot.org/rdf#interactingRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a interacting region annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | InteractingRegionc |
| URI | http://nextprot.org/rdf#interaction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an interaction annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | Interactionc |
| URI | http://nextprot.org/rdf#interactionInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an interaction information annotation |
| Super-properties | interactionop |
| Domain(s) | Isoformc |
| Range(s) | InteractionInfoc |
| URI | http://nextprot.org/rdf#interactionMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an interaction mapping annotation |
| Super-properties | mappingop |
| Domain(s) | Isoformc |
| Range(s) | InteractionMappingc |
| URI | http://nextprot.org/rdf#intramembraneRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to an intramembrane region annotation |
| Super-properties | topologyop |
| Domain(s) | Isoformc |
| Range(s) | IntramembraneRegionc |
| URI | http://nextprot.org/rdf#isoform |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein entry to an isoform |
| Domain(s) | Proteinc |
| Range(s) | Isoformc |
| URI | http://nextprot.org/rdf#kineticKM |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a kinetic KM annotation |
| Super-properties | biophysicochemical propertyop |
| Domain(s) | Isoformc |
| Range(s) | KineticKMc |
| URI | http://nextprot.org/rdf#kineticNote |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a kinetic note annotation |
| Super-properties | biophysicochemical propertyop |
| Domain(s) | Isoformc |
| Range(s) | KineticNotec |
| URI | http://nextprot.org/rdf#kineticVmax |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a kinetic Vmax annotation |
| Super-properties | biophysicochemical propertyop |
| Domain(s) | Isoformc |
| Range(s) | KineticVmaxc |
| URI | http://nextprot.org/rdf#lipidationSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a lipid moiety-binding region annotation |
| Super-properties | ptmop |
| Domain(s) | Isoformc |
| Range(s) | LipidationSitec |
| URI | http://nextprot.org/rdf#lowExpression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a low level of expression |
| Super-properties | detected expressionop |
| Domain(s) | Isoformc |
| Range(s) | ExpressionProfilec |
| URI | http://nextprot.org/rdf#mapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a mapping annotation (antibody mapping, peptide mapping, etc.) |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | Mappingc |
| URI | http://nextprot.org/rdf#matureProtein |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a mature protein annotation |
| Super-properties | processing productop |
| Domain(s) | Isoformc |
| Range(s) | MatureProteinc |
| URI | http://nextprot.org/rdf#medical |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a medical annotation (disease, pharmaceutical, etc.) |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | Medicalc |
| URI | http://nextprot.org/rdf#mediumExpression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a medium level of expression |
| Super-properties | detected expressionop |
| Domain(s) | Isoformc |
| Range(s) | ExpressionProfilec |
| URI | http://nextprot.org/rdf#metalBindingSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a metal binding site annotation |
| Super-properties | siteop |
| Domain(s) | Isoformc |
| Range(s) | MetalBindingSitec |
| URI | http://nextprot.org/rdf#miscellaneous |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a miscellaneous annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | Miscellaneousc |
| URI | http://nextprot.org/rdf#miscellaneousRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a miscellaneous region annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | MiscellaneousRegionc |
| URI | http://nextprot.org/rdf#miscellaneousSite |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a miscellaneous site annotation |
| Super-properties | siteop |
| Domain(s) | Isoformc |
| Range(s) | MiscellaneousSitec |
| URI | http://nextprot.org/rdf#mitochondrialTransitPeptide |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a mitochondrial transit peptide annotation |
| Super-properties | processing productop |
| Domain(s) | Isoformc |
| Range(s) | MitochondrialTransitPeptidec |
| URI | http://nextprot.org/rdf#modifiedResidue |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a modified residue annotation |
| Super-properties | ptmop |
| Domain(s) | Isoformc |
| Range(s) | ModifiedResiduec |
| URI | http://nextprot.org/rdf#mutagenesis |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a mutagenesis annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | Mutagenesisc |
| URI | http://nextprot.org/rdf#name |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Most general property linking a protein, isoform or gene to a name. Also used for person and consortium |
| Domain(s) | (Proteinc or Isoformc or Genec or Personc or Consortiumc) |
| Range(s) | http://nextprot.org/rdf#Name http://www.w3.org/2001/XMLSchema#string |
| URI | http://nextprot.org/rdf#negativeEvidence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an annotation to an evidence that contradicts the annotation statement |
| Domain(s) | Annotationc |
| Range(s) | Evidencec |
| URI | http://nextprot.org/rdf#nonConsecutiveResidue |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a non-consecutive residue annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | NonConsecutiveResiduec |
| URI | http://nextprot.org/rdf#nonTerminalResidue |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a non-terminal residue annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | NonTerminalResiduec |
| URI | http://nextprot.org/rdf#nucleotidePhosphateBindingRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a nucleotide phosphate-binding region annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | NucleotidePhosphateBindingRegionc |
| URI | http://nextprot.org/rdf#observedExpression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence to an observed expression level |
| Domain(s) | Evidencec |
| Range(s) | ObservedExpressionc |
| URI | http://nextprot.org/rdf#orfName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a gene to an ORF name |
| Super-properties | nameop |
| Domain(s) | (Proteinc or Genec) |
| Range(s) | ORFNamec |
| URI | http://nextprot.org/rdf#otherName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links the recommended name of an entity to another name which is sometimes used to refer to the same entity |
| Domain(s) | Namec |
| Range(s) | Namec |
| URI | http://nextprot.org/rdf#pathway |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a pathway annotation |
| Super-properties | functionop |
| Domain(s) | Isoformc |
| Range(s) | Pathwayc |
| URI | http://nextprot.org/rdf#pdbMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a PDB mapping annotation |
| Super-properties | mappingop |
| Domain(s) | Isoformc |
| Range(s) | PdbMappingc |
| URI | http://nextprot.org/rdf#peptideMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a peptide mapping annotation |
| Super-properties | mappingop |
| Domain(s) | Isoformc |
| Range(s) | PeptideMappingc |
| URI | http://nextprot.org/rdf#peroxisomeTransitPeptide |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a peroxisome transit peptide annotation |
| Super-properties | processing productop |
| Domain(s) | Isoformc |
| Range(s) | PeroxisomeTransitPeptidec |
| URI | http://nextprot.org/rdf#phDependence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a pH dependence annotation |
| Super-properties | biophysicochemical propertyop |
| Domain(s) | Isoformc |
| Range(s) | PhDependencec |
| URI | http://nextprot.org/rdf#pharmaceutical |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a pharmaceutical annotation |
| Super-properties | medicalop |
| Domain(s) | Isoformc |
| Range(s) | Pharmaceuticalc |
| URI | http://nextprot.org/rdf#phenotypicVariation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a proteoform to a phenotype variation annotation |
| Super-properties | general annotationop |
| Domain(s) | Proteoformc |
| Range(s) | PhenotypicVariationc |
| URI | http://nextprot.org/rdf#positionalAnnotation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a positional annotation |
| Domain(s) | Isoformc |
| Range(s) | PositionalAnnotationc |
| URI | http://nextprot.org/rdf#processingProduct |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a processing product annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | ProcessingProductc |
| URI | http://nextprot.org/rdf#propeptide |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a maturation peptide annotation |
| Super-properties | processing productop |
| Domain(s) | Isoformc |
| Range(s) | Propeptidec |
| URI | http://nextprot.org/rdf#proteoform |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a proteoform |
| Domain(s) | Isoformc |
| Range(s) | Proteoformc |
| URI | http://nextprot.org/rdf#provenance |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a cross-reference or identifier to its database |
| Domain(s) | (Cross referencec or Identifierc) |
| Range(s) | Databasec |
| URI | http://nextprot.org/rdf#ptm |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a post-translational modification (PTM) annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | Ptmc |
| URI | http://nextprot.org/rdf#ptmInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a post-translational modification (PTM) information annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | PtmInfoc |
| URI | http://nextprot.org/rdf#quality |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an annotation or evidence to a quality qualifier |
| Domain(s) | (Annotationc or Evidencec) |
| Range(s) | QualityQualifierc |
| URI | http://nextprot.org/rdf#recommendedName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein entry, isoform or gene to a recommended name |
| Super-properties | nameop |
| Domain(s) | (Proteinc or Isoformc or Genec) |
| Range(s) | Namec |
| URI | http://nextprot.org/rdf#redoxPotential |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a redox potential annotation |
| Super-properties | biophysicochemical propertyop |
| Domain(s) | Isoformc |
| Range(s) | RedoxPotentialc |
| URI | http://nextprot.org/rdf#reference |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an entry or an evidence to an identifier, a publication or a cross-reference |
| Domain(s) | (Evidencec or Proteinc) |
| Range(s) | http://nextprot.org/rdf#Xref http://nextprot.org/rdf#Identifier http://nextprot.org/rdf#Publication |
| URI | http://nextprot.org/rdf#region |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a region annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | Regionc |
| URI | http://nextprot.org/rdf#repeat |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a repeat annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | Repeatc |
| URI | http://nextprot.org/rdf#secondaryStructure |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a secondary structure annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | SecondaryStructurec |
| URI | http://nextprot.org/rdf#selenocysteine |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a selenocysteine annotation |
| Super-properties | ptmop |
| Domain(s) | Isoformc |
| Range(s) | Selenocysteinec |
| URI | http://nextprot.org/rdf#sequence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a sequence |
| Domain(s) | Isoformc |
| Range(s) | ProteinSequencec |
| URI | http://nextprot.org/rdf#sequenceCaution |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a sequence caution annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | SequenceCautionc |
| URI | http://nextprot.org/rdf#sequenceConflict |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a sequence conflict annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | SequenceConflictc |
| URI | http://nextprot.org/rdf#shortSequenceMotif |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a short sequence motif annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | ShortSequenceMotifc |
| URI | http://nextprot.org/rdf#signalPeptide |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a signal peptide annotation |
| Super-properties | processing productop |
| Domain(s) | Isoformc |
| Range(s) | SignalPeptidec |
| URI | http://nextprot.org/rdf#site |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a site annotation (active site, binding site, etc.) |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | Sitec |
| URI | http://nextprot.org/rdf#smallMoleculeInteraction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a small molecule interaction annotation |
| Super-properties | interactionop |
| Domain(s) | Isoformc |
| Range(s) | SmallMoleculeInteractionc |
| URI | http://nextprot.org/rdf#srmPeptideMapping |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a SRM peptide mapping annotation |
| Super-properties | mappingop |
| Domain(s) | Isoformc |
| Range(s) | SrmPeptideMappingc |
| URI | http://nextprot.org/rdf#subcellularLocation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a subcellular location annotation |
| Super-properties | cellular componentop |
| Domain(s) | Isoformc |
| Range(s) | SubcellularLocationc |
| URI | http://nextprot.org/rdf#subcellularLocationNote |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a subcellular location information annotation |
| Super-properties | cellular componentop |
| Domain(s) | Isoformc |
| Range(s) | SubcellularLocationNotec |
| URI | http://nextprot.org/rdf#temperatureDependence |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a temperature dependence annotation |
| Super-properties | biophysicochemical propertyop |
| Domain(s) | Isoformc |
| Range(s) | TemperatureDependencec |
| URI | http://nextprot.org/rdf#term |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an annotation to a term |
| Domain(s) | (General Annotationc or Positional annotationc) |
| Range(s) | Termc |
| URI | http://nextprot.org/rdf#tissue |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an experimental context to a tissue |
| Domain(s) | Experimental contextc |
| Range(s) | NextprotAnatomyCvc |
| URI | http://nextprot.org/rdf#topologicalDomain |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a topological domain annotation |
| Super-properties | topologyop |
| Domain(s) | Isoformc |
| Range(s) | TopologicalDomainc |
| URI | http://nextprot.org/rdf#topology |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a topology annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | Topologyc |
| URI | http://nextprot.org/rdf#transmembraneRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a transmembrane region annotation |
| Super-properties | topologyop |
| Domain(s) | Isoformc |
| Range(s) | TransmembraneRegionc |
| URI | http://nextprot.org/rdf#transportActivity |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a transport activity annotation |
| Super-properties | functionop |
| Domain(s) | Isoformc |
| Range(s) | TransportActivityc |
| URI | http://nextprot.org/rdf#turn |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a turn annotation |
| Super-properties | secondary structureop |
| Domain(s) | Isoformc |
| Range(s) | Turnc |
| URI | http://nextprot.org/rdf#undetectedExpression |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to undetected level of expression in all experiments |
| Super-properties | expressionop |
| Domain(s) | Isoformc |
| Range(s) | ExpressionProfilec |
| URI | http://nextprot.org/rdf#uniprotKeyword |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a UniProt keyword annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | UniprotKeywordc |
| URI | http://nextprot.org/rdf#variant |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a variant annotation |
| Super-properties | positional annotationop |
| Domain(s) | Isoformc |
| Range(s) | Variantc |
| URI | http://nextprot.org/rdf#variantInfo |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a variant information annotation |
| Super-properties | general annotationop |
| Domain(s) | Isoformc |
| Range(s) | VariantInfoc |
| URI | http://nextprot.org/rdf#zincFingerRegion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to a zinc finger region annotation |
| Super-properties | regionop |
| Domain(s) | Isoformc |
| Range(s) | ZincFingerRegionc |
| URI | http://nextprot.org/rdf#accession |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a cross-reference or an identifier to a string uniquely identifying it in its source database |
| Domain(s) | (Cross referencec or Identifierc) |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#alleleCount |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence for a variant (allele) to its count in a population |
| Domain(s) | Evidencec |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#alleleFrequency |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence for a variant (allele) to its frequency in a population |
| Domain(s) | Evidencec |
| Range(s) | xsd:doublec |
| URI | http://nextprot.org/rdf#alleleNumber |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence for a variant (allele) to the total number of alleles assessed in a population |
| Domain(s) | Evidencec |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#antibodyName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an antibody mapping annotation to the antibody name |
| Domain(s) | Antibody mappingc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#antibodyUniqueness |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an antibody mapping annotation to its unicity, i.e. whether it maps uniquely to a protein entry or not |
| Domain(s) | Antibody mappingc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#band |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a gene to a chromosomal band |
| Domain(s) | Genec |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#begin |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a gene to the position on the chromosome marking the beginning of the gene |
| Domain(s) | Genec |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#category |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a database to its category |
| Domain(s) | Databasec |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#chain |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a sequence to a chain |
| Domain(s) | Protein sequencec |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#chromosome |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a gene to a chromosome |
| Domain(s) | Genec |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#city |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to a city where it was published |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#end |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a gene to the position on the chromosome marking the end of the gene or a positional annotation with the end of the positional feature |
| Domain(s) | (Positional annotationc or Genec) |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#entryAnnotationId |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Unique link between an entry and an annotation |
| Domain(s) | (General Annotationc or Positional annotationc) |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#expressionScore |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence to an expression score |
| Domain(s) | Evidencec |
| Range(s) | xsd:doublec |
| URI | http://nextprot.org/rdf#firstPage |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to the first page number |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#from |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to a PMID or DOI cross reference |
| Domain(s) | Publicationc |
| Range(s) | Xrefc |
| URI | http://nextprot.org/rdf#goQualifier |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a GO qualifier to a GO biological process, cellular component or molecular function annotation |
| Domain(s) | Evidencec |
| Range(s) | GoQualifierc |
| URI | http://nextprot.org/rdf#hgvs |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a variant or a mutagenesis annotation to a HGVS name |
| Domain(s) | (Mutagenesisc or Variantc) |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#homozygoteCount |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence for a variant (allele) to its homozygote count in a population |
| Domain(s) | Evidencec |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#integrated |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a UniprotHistory or NextprotHistory to the date of integration |
| Domain(s) | Protein record historyc |
| Range(s) | xsd:datec |
| URI | http://nextprot.org/rdf#interactionDetectionMethod |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence for a binary interaction to an interaction detection method |
| Domain(s) | Evidencec |
| Range(s) | PsiMiCvc |
| URI | http://nextprot.org/rdf#isoelectricPoint |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform sequence to an isoelectric point |
| Domain(s) | Protein sequencec |
| Range(s) | xsd:doublec |
| URI | http://nextprot.org/rdf#isoformCount |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein entry to the isoform count |
| Domain(s) | Proteinc |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#issue |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to an issue number |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#journal |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to a journal |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#lastPage |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to the last page number |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#lastSequenceUpdate |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a UniprotHistory to the last sequence update date |
| Domain(s) | UniProt historyc |
| Range(s) | xsd:datec |
| URI | http://nextprot.org/rdf#length |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform sequence to a length (no. of amino acids) |
| Domain(s) | (Genec or Protein sequencec) |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#level |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein existence annotation to its numerical value |
| Domain(s) | Protein existencec |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#metadata |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence to metadata |
| Domain(s) | Experimental contextc |
| Range(s) | Publicationc |
| URI | http://nextprot.org/rdf#method |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a pdb mapping annotation to a method |
| Domain(s) | PDB mappingc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#molecularWeight |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform sequence to a molecular weight |
| Domain(s) | Protein sequencec |
| Range(s) | xsd:doublec |
| URI | http://nextprot.org/rdf#negative |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
If the value is True,then the evidence contradicts the annotation statement |
| Domain(s) | Evidencec |
| Range(s) | xsd:booleanc |
| URI | http://nextprot.org/rdf#numberOfExperiments |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an evidence of a binary interaction annotation to the number of experiments |
| Domain(s) | Evidencec |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#original |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a variant, mutagenesis or conflict annotation to the original amino acid(s) found at the position of the variant |
| Domain(s) | (Sequence conflictc or Mutagenesisc or Variantc) |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#peptideName |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a peptide mapping or a SRM peptide mapping annotation to the name of the peptide or SRM peptide |
| Domain(s) | (SRM peptide mappingc or Peptide mappingc) |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#peptideSource |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a peptide mapping annotation to its source (database or publication) |
| Domain(s) | Peptide mappingc |
| Range(s) | Sourcec |
| URI | http://nextprot.org/rdf#peptideUniqueness |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a peptide mapping annotation or a SRM peptide mapping annotation to its uniqueness, i.e. whether it maps uniquely to a protein entry or not |
| Domain(s) | (SRM peptide mappingc or Peptide mappingc) |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#proteotypic |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a peptide or SRM peptide mapping annotation to a proteotypic flag, i.e. whether it maps uniquely or pseudo-uniquely to a protein entry or not |
| Domain(s) | (SRM peptide mappingc or Peptide mappingc) |
| Range(s) | xsd:booleanc |
| URI | http://nextprot.org/rdf#pubType |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to a publication type (article, book, etc.) |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#publisher |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to a publisher |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#selfInteraction |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a binary interaction annotation to a self interaction value, i.e. flag indicating that a protein entry or isoform interacts with itself |
| Domain(s) | Binary interactionc |
| Range(s) | xsd:booleanc |
| URI | http://nextprot.org/rdf#sequenceVersion |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a Uniprot history to a version number |
| Domain(s) | UniProt historyc |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#severity |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links the evidence of a phenotypic variation annotation to a severity |
| Domain(s) | Evidencec |
| Range(s) | Severityc |
| URI | http://nextprot.org/rdf#start |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a positional annotation with the start of the positional feature |
| Domain(s) | Positional annotationc |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#strand |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a gene to a chromosome strand |
| Domain(s) | Genec |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#suffix |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a person to a suffix in their name |
| Domain(s) | Personc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#swissprotDisplayed |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links an isoform to it being displayed as canonical in UniProt/Swiss-Prot |
| Domain(s) | Isoformc |
| Range(s) | xsd:booleanc |
| URI | http://nextprot.org/rdf#swissprotPage |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a protein entry to a UniProt/Swiss-Prot protein entry web page |
| Domain(s) | Proteinc |
| Range(s) | rdfs:Resourcec |
| URI | http://nextprot.org/rdf#title |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to a title |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#updated |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a UniprotHistory or NextprotHistory to the last update date |
| Domain(s) | Protein record historyc |
| Range(s) | xsd:datec |
| URI | http://nextprot.org/rdf#variation |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a variant, mutagenesis or conflict annotation to the amino acid(s) variation found at the position of the variant |
| Domain(s) | (Sequence conflictc or Mutagenesisc or Variantc) |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#version |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a UniprotHistory to sequence version |
| Domain(s) | Protein record historyc |
| Range(s) | xsd:integerc |
| URI | http://nextprot.org/rdf#volume |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to a volume number |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI | http://nextprot.org/rdf#year |
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description |
Links a publication to the year of publication |
| Domain(s) | Publicationc |
| Range(s) | xsd:stringc |
| URI |
http://nextprot.org/rdf#BRONZE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Quality qualifier describing unreliable annotation or evidence. Excluded from neXtProt. |
| URI |
http://nextprot.org/rdf#Colocalizes_with
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | GO qualifier used to modify the GO cellular component annotation of a protein when the protein transiently or peripherically associates with an organelle or a complex. |
| URI |
http://nextprot.org/rdf#Contributes_to
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | GO qualifier used to modify the molecular function annotation of a protein which is a member of a complex that is defined as an “irreducible molecular machine” - where a particular molecular function cannot be ascribed to an individual subunit or small set of subunits of a complex. |
| URI |
http://nextprot.org/rdf#Enables
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| URI |
http://nextprot.org/rdf#Evidence_at_protein_level
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Entry whose protein(s) existence is based on evidence at protein level (PE1). |
| URI |
http://nextprot.org/rdf#Evidence_at_transcript_level
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Entry whose protein existence is based on evidence at transcript level (PE2). |
| URI |
http://nextprot.org/rdf#GOLD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Quality qualifier describing highly reliable annotation or evidence. |
| URI |
http://nextprot.org/rdf#High
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Expression of the gene product(s) is high. |
| URI |
http://nextprot.org/rdf#Inferred_from_homology
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Entry whose protein existence is based on homology (PE3). |
| URI |
http://nextprot.org/rdf#Is_active_in
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| URI |
http://nextprot.org/rdf#Located_in
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| URI |
http://nextprot.org/rdf#Low
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Expression of the gene product(s) is low. |
| URI |
http://nextprot.org/rdf#Medium
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Expression of the gene product(s) is medium (neither high, nor low). |
| URI |
http://nextprot.org/rdf#Mild
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| URI |
http://nextprot.org/rdf#Moderate
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| URI |
http://nextprot.org/rdf#Negative
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Expression of the gene product(s) is not detectable. |
| URI |
http://nextprot.org/rdf#Part_of
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| URI |
http://nextprot.org/rdf#Positive
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Expression of the gene product(s) is detectable. |
| URI |
http://nextprot.org/rdf#Predicted
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Entry whose protein existence is based on a prediction (gene model) (PE4). |
| URI |
http://nextprot.org/rdf#SILVER
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Quality qualifier describing reliable annotation or evidence. |
| URI |
http://nextprot.org/rdf#Severe
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| URI |
http://nextprot.org/rdf#Uncertain
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Entry whose protein existence is uncertain (PE5). |
| URI |
http://nextprot.org/rdf/db/ABCD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ABCD curated depository of sequenced antibodies |
| See Also | https://web.expasy.org/abcd |
| URI |
http://nextprot.org/rdf/db/Affymetrix
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Affymetrix microarray clone identifier |
| URI |
http://nextprot.org/rdf/db/Allergome
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Allergome; a platform for allergen knowledge |
| See Also | http://www.allergome.org/ |
| URI |
http://nextprot.org/rdf/db/Antibodypedia
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Antibodypedia a portal for validated antibodies |
| See Also | https://antibodypedia.com/ |
| URI |
http://nextprot.org/rdf/db/BMRB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Biological Magnetic Resonance Data Bank |
| See Also | https://bmrb.io/ |
| URI |
http://nextprot.org/rdf/db/BRENDA
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | BRENDA Comprehensive Enzyme Information System |
| See Also | https://www.brenda-enzymes.org |
| URI |
http://nextprot.org/rdf/db/Bgee
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Bgee database of gene expression across multiple animal species |
| See Also | https://bgee.org |
| URI |
http://nextprot.org/rdf/db/BindingDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | BindingDB database of measured binding affinities |
| See Also | https://www.bindingdb.org/ |
| URI |
http://nextprot.org/rdf/db/BioCyc
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | BioCyc Collection of Pathway/Genome Databases |
| See Also | https://www.biocyc.org/ |
| URI |
http://nextprot.org/rdf/db/BioGRID
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The Biological General Repository for Interaction Datasets (BioGRID) |
| See Also | https://thebiogrid.org/ |
| URI |
http://nextprot.org/rdf/db/BioGRID-ORCS
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | BioGRID ORCS database of CRISPR phenotype screens |
| See Also | https://orcs.thebiogrid.org |
| URI |
http://nextprot.org/rdf/db/BioMuta
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | BioMuta curated single-nucleotide variation and disease association database |
| See Also | https://hive.biochemistry.gwu.edu/tools/biomuta/ |
| URI |
http://nextprot.org/rdf/db/CAZy
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Carbohydrate-Active enZymes |
| See Also | http://www.cazy.org/ |
| URI |
http://nextprot.org/rdf/db/CCDS
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The Consensus CDS (CCDS) project |
| See Also | https://www.ncbi.nlm.nih.gov/CCDS |
| URI |
http://nextprot.org/rdf/db/CDD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Conserved Domains Database |
| See Also | https://www.ncbi.nlm.nih.gov/cdd |
| URI |
http://nextprot.org/rdf/db/CORUM
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | CORUM comprehensive resource of mammalian protein complexes |
| See Also | http://mips.helmholtz-muenchen.de/corum/ |
| URI |
http://nextprot.org/rdf/db/CPTAC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The CPTAC Assay portal |
| See Also | https://assays.cancer.gov/ |
| URI |
http://nextprot.org/rdf/db/CPTC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The CPTC Antibody Portal |
| See Also | https://antibodies.cancer.gov |
| URI |
http://nextprot.org/rdf/db/CTD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Comparative Toxicogenomics Database |
| See Also | https://ctdbase.org/ |
| URI |
http://nextprot.org/rdf/db/CarbonylDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | CarbonylDB database of protein carbonylation sites |
| See Also | http://digbio.missouri.edu/CarbonylDB/ |
| URI |
http://nextprot.org/rdf/db/ChEBI
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Chemical Entities of Biological Interest |
| See Also | http://www.ebi.ac.uk/chebi/ |
| URI |
http://nextprot.org/rdf/db/ChEMBL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ChEMBL database of bioactive drug-like small molecules |
| See Also | https://www.ebi.ac.uk/chembldb |
| URI |
http://nextprot.org/rdf/db/ChiTaRS
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data |
| See Also | http://chitars.md.biu.ac.il/ |
| URI |
http://nextprot.org/rdf/db/ComplexPortal
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ComplexPortal: manually curated resource of macromolecular complexes |
| See Also | https://www.ebi.ac.uk/complexportal/ |
| URI |
http://nextprot.org/rdf/db/Cosmic
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | COSMIC, the Catalogue Of Somatic Mutations In Cancer |
| See Also | http://www.sanger.ac.uk/perl/genetics/CGP/core_line_viewer?action=cell_lines |
| URI |
http://nextprot.org/rdf/db/DDBJ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | DNA Data Bank of Japan; a nucleotide sequence database |
| See Also | https://www.ddbj.nig.ac.jp/index-e.html |
| URI |
http://nextprot.org/rdf/db/DECIPHER
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | DECIPHER - Mapping the clinical genome |
| See Also | https://www.deciphergenomics.org/ |
| URI |
http://nextprot.org/rdf/db/DEPOD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | DEPOD human dephosphorylation database |
| See Also | http://depod.bioss.uni-freiburg.de |
| URI |
http://nextprot.org/rdf/db/DIP
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Database of interacting proteins |
| See Also | https://dip.doe-mbi.ucla.edu/ |
| URI |
http://nextprot.org/rdf/db/DKFZ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | DKFZ (Deutsches Krebsforschungszentrum) identifier |
| URI |
http://nextprot.org/rdf/db/DMDM
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Domain mapping of disease mutations (DMDM) |
| See Also | http://bioinf.umbc.edu/dmdm/ |
| URI |
http://nextprot.org/rdf/db/DNASU
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The DNASU plasmid repository |
| See Also | https://dnasu.org/DNASU/ |
| URI |
http://nextprot.org/rdf/db/DOI
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Digital Object Identifier |
| See Also | http://dx.doi.org/ |
| URI |
http://nextprot.org/rdf/db/DOSAC-COBS-2DPAGE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | DOSAC-COBS 2D-PAGE database |
| See Also | http://www.dosac.unipa.it/2d/ |
| URI |
http://nextprot.org/rdf/db/DYP
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The Kahn Dynamic Proteomics Database |
| See Also | http://www.weizmann.ac.il/mcb/UriAlon/DynamProt/ |
| URI |
http://nextprot.org/rdf/db/DisGeNET
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | DisGeNET, a database of gene-disease associations |
| See Also | https://www.disgenet.org/ |
| URI |
http://nextprot.org/rdf/db/DisProt
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Database of protein disorder |
| See Also | https://disprot.org |
| URI |
http://nextprot.org/rdf/db/DrugBank
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Drug and drug target database |
| See Also | https://www.drugbank.ca/ |
| URI |
http://nextprot.org/rdf/db/DrugCentral
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | DrugCentral |
| See Also | https://drugcentral.org/ |
| URI |
http://nextprot.org/rdf/db/ELM
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The Eukaryotic Linear Motif resource for Functional Sites in Proteins |
| See Also | http://elm.eu.org/ |
| URI |
http://nextprot.org/rdf/db/EMBL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | EMBL nucleotide sequence database |
| See Also | https://www.ebi.ac.uk/ena |
| URI |
http://nextprot.org/rdf/db/EMBL-CDS
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | EMBL coding sequence (CDS) database |
| See Also | http://www.ebi.ac.uk/embl/ |
| URI |
http://nextprot.org/rdf/db/EPD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Encyclopedia of Proteome Dynamics |
| See Also | https://www.peptracker.com/epd/analytics/ |
| URI |
http://nextprot.org/rdf/db/ESTHER
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ESTHER database of the Alpha/Beta-hydrolase fold superfamily of proteins |
| See Also | http://bioweb.supagro.inra.fr/ESTHER/general?what=index |
| URI |
http://nextprot.org/rdf/db/Ensembl
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Ensembl eukaryotic genome annotation project |
| See Also | https://www.ensembl.org/ |
| URI |
http://nextprot.org/rdf/db/EvolutionaryTrace
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Relative evolutionary importance of amino acids within a protein sequence |
| See Also | http://lichtargelab.org/software/ETserver |
| URI |
http://nextprot.org/rdf/db/ExpressionAtlas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ExpressionAtlas, Differential and Baseline Expression |
| See Also | https://www.ebi.ac.uk/gxa |
| URI |
http://nextprot.org/rdf/db/FLJ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | FLJ identifier |
| URI |
http://nextprot.org/rdf/db/GFP-cDNA
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | EMBL/DKFZ GFP-cDNA Localisation Project |
| See Also | http://gfp-cdna.embl.de |
| URI |
http://nextprot.org/rdf/db/GOA
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Gene Ontologie Annotation |
| See Also | http://www.ebi.ac.uk/GOA/ |
| URI |
http://nextprot.org/rdf/db/GO_REF
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Gene Ontologie References |
| See Also | http://www.geneontology.org/cgi-bin/references.cgi |
| URI |
http://nextprot.org/rdf/db/Gene3D
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Gene3D Structural and Functional Annotation of Protein Families |
| See Also | http://gene3d.biochem.ucl.ac.uk/Gene3D/ |
| URI |
http://nextprot.org/rdf/db/GeneCards
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | GeneCards: human genes, protein and diseases |
| See Also | https://www.genecards.org/ |
| URI |
http://nextprot.org/rdf/db/GeneDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | GeneDB pathogen genome database from Sanger Institute |
| See Also | https://www.genedb.org/ |
| URI |
http://nextprot.org/rdf/db/GeneID
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Database of genes from NCBI RefSeq genomes |
| See Also | https://www.ncbi.nlm.nih.gov/gene |
| URI |
http://nextprot.org/rdf/db/GeneReviews
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | GeneReviews a resource of expert-authored, peer-reviewed disease descriptions. |
| See Also | https://www.ncbi.nlm.nih.gov/books/NBK1116 |
| URI |
http://nextprot.org/rdf/db/GeneTree
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Ensembl GeneTree |
| See Also | https://ensemblgenomes.org |
| URI |
http://nextprot.org/rdf/db/GeneWiki
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The Gene Wiki collection of pages on human genes and proteins |
| See Also | https://en.wikipedia.org/wiki/Portal:Gene_Wiki |
| URI |
http://nextprot.org/rdf/db/Genevisible
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Genevisible search portal to normalized and curated expression data from Genevestigator |
| See Also | https://genevisible.com/search |
| URI |
http://nextprot.org/rdf/db/GenomeRNAi
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Database of phenotypes from RNA interference screens in Drosophila and Homo sapiens |
| See Also | http://genomernai.dkfz.de/ |
| URI |
http://nextprot.org/rdf/db/GlyConnect
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | GlyConnect protein glycosylation platform |
| See Also | https://glyconnect.expasy.org |
| URI |
http://nextprot.org/rdf/db/GlyGen
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | GlyGen: Computational and Informatics Resources for Glycoscience |
| See Also | https://glygen.org |
| URI |
http://nextprot.org/rdf/db/GuidetoPHARMACOLOGY
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | IUPHAR/BPS Guide to PHARMACOLOGY |
| See Also | https://www.guidetopharmacology.org |
| URI |
http://nextprot.org/rdf/db/HAMAP
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | HAMAP database of protein families |
| See Also | https://hamap.expasy.org/ |
| URI |
http://nextprot.org/rdf/db/HAMAP-Rule
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | High-quality Automated and Manual Annotation of Proteins |
| See Also | https://hamap.expasy.org/ |
| URI |
http://nextprot.org/rdf/db/HGNC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | HUGO Gene Nomenclature Committee identifier |
| See Also | https://www.genenames.org/ |
| URI |
http://nextprot.org/rdf/db/HOGENOM
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The HOGENOM Database of Homologous Genes from Fully Sequenced Organisms |
| See Also | http://hogenom.univ-lyon1.fr/ |
| URI |
http://nextprot.org/rdf/db/HPA
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Human Protein Atlas |
| See Also | https://www.proteinatlas.org/ |
| URI |
http://nextprot.org/rdf/db/HPRD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Human Proteins Reference Database |
| See Also | http://www.hprd.org |
| URI |
http://nextprot.org/rdf/db/IDEAL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Intrinsically Disordered proteins with Extensive Annotations and Literature |
| See Also | http://idp1.force.cs.is.nagoya-u.ac.jp/IDEAL/ |
| URI |
http://nextprot.org/rdf/db/IMAGE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | IMAGE Consortium clone identifier |
| URI |
http://nextprot.org/rdf/db/IMGT_GENE-DB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The international ImMunoGeneTics information system |
| See Also | http://www.imgt.org/genedb |
| URI |
http://nextprot.org/rdf/db/IPI
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | International Protein Index |
| See Also | http://www.ebi.ac.uk/IPI |
| URI |
http://nextprot.org/rdf/db/Illumina
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Illumina microarray clone identifier |
| URI |
http://nextprot.org/rdf/db/InParanoid
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | InParanoid: Eukaryotic Ortholog Groups |
| See Also | https://inparanoid.sbc.su.se/ |
| URI |
http://nextprot.org/rdf/db/IntAct
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | IntAct Molecular Interaction Database |
| See Also | https://www.ebi.ac.uk/intact/ |
| URI |
http://nextprot.org/rdf/db/InterPro
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Integrated resource of protein families, domains and functional sites |
| See Also | https://www.ebi.ac.uk/interpro/ |
| URI |
http://nextprot.org/rdf/db/KEGG
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | KEGG: Kyoto Encyclopedia of Genes and Genomes |
| See Also | https://www.genome.jp/kegg/ |
| URI |
http://nextprot.org/rdf/db/KEGGPathway
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | KEGG Pathway is a collection of manually drawn pathway maps |
| See Also | http://www.genome.jp/kegg/pathway.html |
| URI |
http://nextprot.org/rdf/db/KIAA
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Identifier subtype |
| URI |
http://nextprot.org/rdf/db/LOC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Entrez Gene symbol |
| See Also | http://www.ncbi.nlm.nih.gov/gene |
| URI |
http://nextprot.org/rdf/db/MEROPS
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | MEROPS protease database |
| See Also | https://www.ebi.ac.uk/merops/ |
| URI |
http://nextprot.org/rdf/db/MGC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Identifier subtype |
| URI |
http://nextprot.org/rdf/db/MGI
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Mouse genome database (MGD) from Mouse Genome Informatics (MGI) |
| See Also | http://www.informatics.jax.org/ |
| URI |
http://nextprot.org/rdf/db/MIM
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Online Mendelian Inheritance in Man (OMIM) |
| See Also | https://www.omim.org/ |
| URI |
http://nextprot.org/rdf/db/MINT
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Molecular INTeraction database |
| See Also | https://mint.bio.uniroma2.it/ |
| URI |
http://nextprot.org/rdf/db/MalaCards
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | MalaCards human disease database |
| See Also | https://www.malacards.org |
| URI |
http://nextprot.org/rdf/db/MassIVE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | MassIVE - Mass Spectrometry Interactive Virtual Environment |
| See Also | https://massive.ucsd.edu/ |
| URI |
http://nextprot.org/rdf/db/MaxQB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | MaxQB - The MaxQuant DataBase |
| See Also | http://maxqb.biochem.mpg.de/mxdb/ |
| URI |
http://nextprot.org/rdf/db/MetOSite
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | MetOSite database of methionine sulfoxide sites |
| See Also | https://metosite.uma.es/ |
| URI |
http://nextprot.org/rdf/db/MoonDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | MoonDB Database of extreme multifunctional and moonlighting proteins |
| See Also | http://moondb.hb.univ-amu.fr |
| URI |
http://nextprot.org/rdf/db/MoonProt
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | MoonProt database of moonlighting proteins |
| See Also | http://www.moonlightingproteins.org/ |
| URI |
http://nextprot.org/rdf/db/NCBI_Taxonomy
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | NCBI taxonomy database |
| See Also | http://www.ncbi.nlm.nih.gov/Taxonomy |
| URI |
http://nextprot.org/rdf/db/NIAGADS
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | NIAGADS Genomics Database |
| See Also | https://www.niagads.org/genomics/ |
| URI |
http://nextprot.org/rdf/db/OGP
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | USC-OGP 2-DE database |
| See Also | http://usc_ogp_2ddatabase.cesga.es/cgi-bin/2d/2d.cgi |
| URI |
http://nextprot.org/rdf/db/OMA
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Identification of Orthologs from Complete Genome Data |
| See Also | https://omabrowser.org/ |
| URI |
http://nextprot.org/rdf/db/OpenTargets
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Open Targets |
| See Also | https://platform.opentargets.org/ |
| URI |
http://nextprot.org/rdf/db/Orphanet
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Orphanet; a database dedicated to information on rare diseases and orphan drugs |
| See Also | https://www.orpha.net/consor/cgi-bin/index.php |
| URI |
http://nextprot.org/rdf/db/OrthoDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Database of Orthologous Groups |
| See Also | https://www.orthodb.org |
| URI |
http://nextprot.org/rdf/db/Others
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Identifier from another source |
| URI |
http://nextprot.org/rdf/db/PANTHER
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The PANTHER Classification System |
| See Also | http://www.pantherdb.org/ |
| URI |
http://nextprot.org/rdf/db/PATRIC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Pathosystems Resource Integration Center (PATRIC) |
| See Also | https://patricbrc.org/ |
| URI |
http://nextprot.org/rdf/db/PCDDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The Protein Circular Dichroism Data Bank |
| See Also | https://pcddb.cryst.bbk.ac.uk/ |
| URI |
http://nextprot.org/rdf/db/PDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Protein Data Bank Europe |
| See Also | https://www.ebi.ac.uk/pdbe/ |
| URI |
http://nextprot.org/rdf/db/PDBsum
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PDBsum; at-a-glance overview of macromolecular structures |
| See Also | https://www.ebi.ac.uk/pdbsum/ |
| URI |
http://nextprot.org/rdf/db/PIR
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Protein Information Resource (PIR) identifier |
| See Also | https://proteininformationresource.org/ |
| URI |
http://nextprot.org/rdf/db/PIRNR
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PIR Name Rule |
| See Also | http://www.uniprot.org/ |
| URI |
http://nextprot.org/rdf/db/PIRSF
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PIRSF; a whole-protein classification database |
| See Also | https://proteininformationresource.org/pirwww/dbinfo/pirsf.shtml |
| URI |
http://nextprot.org/rdf/db/PIRSR
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PIRSF site rule system |
| See Also | http://pir.georgetown.edu |
| URI |
http://nextprot.org/rdf/db/PRIDE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PRoteomics IDEntifications database |
| See Also | https://www.ebi.ac.uk/pride |
| URI |
http://nextprot.org/rdf/db/PRINTS
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Protein Motif fingerprint database; a protein domain database |
| See Also | http://www.bioinf.manchester.ac.uk/dbbrowser/PRINTS/ |
| URI |
http://nextprot.org/rdf/db/PRO
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Protein Ontology |
| See Also | https://proconsortium.org/ |
| URI |
http://nextprot.org/rdf/db/PROSITE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PROSITE; a protein domain and family database |
| See Also | https://prosite.expasy.org/ |
| URI |
http://nextprot.org/rdf/db/PROSITE-ProRule
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ProRule section of PROSITE |
| See Also | https://prosite.expasy.org/ |
| URI |
http://nextprot.org/rdf/db/PathwayCommons
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Pathway Commons web resource for biological pathway data |
| See Also | http://www.pathwaycommons.org |
| URI |
http://nextprot.org/rdf/db/PaxDb
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PaxDb, a database of protein abundance averages across all three domains of life |
| See Also | https://pax-db.org |
| URI |
http://nextprot.org/rdf/db/PeptideAtlas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PeptideAtlas |
| See Also | http://www.peptideatlas.org |
| URI |
http://nextprot.org/rdf/db/PeroxiBase
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PeroxiBase, a peroxidase database |
| See Also | http://peroxibase.toulouse.inra.fr/ |
| URI |
http://nextprot.org/rdf/db/Pfam
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Pfam protein domain database |
| See Also | https://pfam.xfam.org/ |
| URI |
http://nextprot.org/rdf/db/PharmGKB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The Pharmacogenetics and Pharmacogenomics Knowledge Base |
| See Also | https://www.pharmgkb.org/ |
| URI |
http://nextprot.org/rdf/db/Pharos
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Pharos NIH Druggable Genome Knowledgebase |
| See Also | https://pharos.nih.gov |
| URI |
http://nextprot.org/rdf/db/PhosphoSitePlus
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Comprehensive resource for the study of protein post-translational modifications (PTMs) in human, mouse and rat. |
| See Also | https://www.phosphosite.org |
| URI |
http://nextprot.org/rdf/db/PhylomeDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Database for complete collections of gene phylogenies |
| See Also | http://phylomedb.org/ |
| URI |
http://nextprot.org/rdf/db/Proteomes
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Proteomes |
| See Also | https://www.uniprot.org/proteomes |
| URI |
http://nextprot.org/rdf/db/ProteomicsDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ProteomicsDB: a multi-organism proteome resource |
| See Also | https://www.proteomicsdb.org/ |
| URI |
http://nextprot.org/rdf/db/PubMed
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | PubMed site |
| See Also | http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed |
| URI |
http://nextprot.org/rdf/db/REBASE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Restriction enzymes and methylases database |
| See Also | http://rebase.neb.com/rebase/rebase.html |
| URI |
http://nextprot.org/rdf/db/REPRODUCTION-2DPAGE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | REPRODUCTION-2DPAGE |
| See Also | http://reprod.njmu.edu.cn/cgi-bin/2d/2d.cgi |
| URI |
http://nextprot.org/rdf/db/RNAct
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | RNAct, Protein-RNA interaction predictions for model organisms. |
| See Also | https://rnact.crg.eu |
| URI |
http://nextprot.org/rdf/db/Reactome
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Reactome - a knowledgebase of biological pathways and processes |
| See Also | https://reactome.org |
| URI |
http://nextprot.org/rdf/db/RefSeq
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | NCBI Reference Sequences |
| See Also | https://www.ncbi.nlm.nih.gov/refseq/ |
| URI |
http://nextprot.org/rdf/db/Rhea
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Rhea - the annotated database of biochemical reactions |
| See Also | https://www.rhea-db.org/ |
| URI |
http://nextprot.org/rdf/db/RuleBase
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | UniProtKB annotation rules |
| See Also | https://services.uniprot.org/supplement/ |
| URI |
http://nextprot.org/rdf/db/SABIO-RK
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | SABIO-RK: Biochemical Reaction Kinetics Database |
| See Also | http://sabiork.h-its.org/ |
| URI |
http://nextprot.org/rdf/db/SAM
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | SAM - Sequence Analysis Methods for automatic annotation |
| See Also | https://mobidb.bio.unipd.it/ |
| URI |
http://nextprot.org/rdf/db/SASBDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Small Angle Scattering Biological Data Bank |
| See Also | https://www.sasbdb.org/ |
| URI |
http://nextprot.org/rdf/db/SFLD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Structure-Function Linkage Database |
| See Also | http://sfld.rbvi.ucsf.edu/django/ |
| URI |
http://nextprot.org/rdf/db/SIGNOR
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | SIGNOR Signaling Network Open Resource |
| See Also | https://signor.uniroma2.it/ |
| URI |
http://nextprot.org/rdf/db/SMART
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Simple Modular Architecture Research Tool; a protein domain database |
| See Also | http://smart.embl.de/ |
| URI |
http://nextprot.org/rdf/db/SMR
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | SWISS-MODEL Repository - a database of annotated 3D protein structure models |
| See Also | https://swissmodel.expasy.org/repository/ |
| URI |
http://nextprot.org/rdf/db/SRMAtlas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | SRMAtlas |
| See Also | http://www.srmatlas.org |
| URI |
http://nextprot.org/rdf/db/STRING
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | STRING: functional protein association networks |
| See Also | https://string-db.org/ |
| URI |
http://nextprot.org/rdf/db/SUPFAM
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Superfamily database of structural and functional annotation |
| See Also | https://supfam.org |
| URI |
http://nextprot.org/rdf/db/SWISS-2DPAGE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Two-dimensional polyacrylamide gel electrophoresis database from the Geneva University Hospital |
| See Also | https://world-2dpage.expasy.org/swiss-2dpage/ |
| URI |
http://nextprot.org/rdf/db/SignaLink
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | SignaLink: a signaling pathway resource with multi-layered regulatory networks |
| See Also | http://signalink.org/ |
| URI |
http://nextprot.org/rdf/db/SwissLipids
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | SwissLipids knowledge resource for lipid biology |
| See Also | https://www.swisslipids.org |
| URI |
http://nextprot.org/rdf/db/SwissPalm
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | SwissPalm database of S-palmitoylation events |
| See Also | https://swisspalm.org |
| URI |
http://nextprot.org/rdf/db/TCDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Transport Classification Database |
| See Also | http://www.tcdb.org/ |
| URI |
http://nextprot.org/rdf/db/TIGRFAMs
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | TIGRFAMs; a protein family database |
| See Also | https://www.ncbi.nlm.nih.gov/genome/annotation_prok/tigrfams/ |
| URI |
http://nextprot.org/rdf/db/TopDownProteomics
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Consortium for Top Down Proteomics |
| See Also | http://repository.topdownproteomics.org/ |
| URI |
http://nextprot.org/rdf/db/TreeFam
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | TreeFam database of animal gene trees |
| See Also | http://www.treefam.org |
| URI |
http://nextprot.org/rdf/db/UCD-2DPAGE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | University College Dublin 2-DE Proteome Database |
| See Also | https://proteomics-portal.ucd.ie/cgi-bin/2d/2d.cgi |
| URI |
http://nextprot.org/rdf/db/UCSC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | UCSC genome browser |
| See Also | https://genome.ucsc.edu/ |
| URI |
http://nextprot.org/rdf/db/UniLectin
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | UniLectin database of carbohydrate-binding proteins |
| See Also | https://unilectin.eu |
| URI |
http://nextprot.org/rdf/db/UniPathway
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | UniPathway: a resource for the exploration and annotation of metabolic pathways |
| See Also | http://www.unipathway.org |
| URI |
http://nextprot.org/rdf/db/UniProt
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | UniProt web site |
| See Also | http://www.uniprot.org/uniprot/ |
| URI |
http://nextprot.org/rdf/db/UniProtKB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Identifier subtype |
| URI |
http://nextprot.org/rdf/db/VEuPathDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Eukaryotic Pathogen, Vector and Host Database Resources |
| See Also | https://veupathdb.org/veupathdb/app |
| URI |
http://nextprot.org/rdf/db/dbSNP
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Database of single nucleotide polymorphism |
| See Also | https://www.ncbi.nlm.nih.gov/SNP/ |
| URI |
http://nextprot.org/rdf/db/eggNOG
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | evolutionary genealogy of genes: Non-supervised Orthologous Groups |
| See Also | http://eggnogdb.embl.de/ |
| URI |
http://nextprot.org/rdf/db/gnomAD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Genome aggregation database |
| See Also | https://gnomad.broadinstitute.org/ |
| URI |
http://nextprot.org/rdf/db/iPTMnet
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | iPTMnet integrated resource for PTMs in systems biology context |
| See Also | https://research.bioinformatics.udel.edu/iptmnet/ |
| URI |
http://nextprot.org/rdf/db/jPOST
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | jPOST - Japan Proteome Standard Repository/Database |
| See Also | https://globe.jpostdb.org/ |
| URI |
http://nextprot.org/rdf/db/neXtProt
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | neXtProt; the human protein knowledge platform |
| See Also | https://www.nextprot.org/ |
| URI |
http://nextprot.org/rdf/db/neXtProtSubmission
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | neXtProt submission; the human protein knowledge platform, publication of type submission part |
| See Also | http://www.nextprot.org/ |
| URI |
http://nextprot.org/rdf/source/ARUK-UCL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Alzheimers Research Gene Ontology Initiative (Alzheimer's research UK). |
| See Also | http://www.ucl.ac.uk/functional-gene-annotation/neurological |
| URI |
http://nextprot.org/rdf/source/AgBase
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | AgBase is a curated resource for functional analysis of agricultural plant and animal gene products. |
| See Also | http://agbase.msstate.edu/ |
| URI |
http://nextprot.org/rdf/source/Alzheimers_University_of_Toronto
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Alzheimers Project at University of Toronto |
| See Also | http://www.ims.utoronto.ca/Page4.aspx |
| URI |
http://nextprot.org/rdf/source/BHF-UCL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | British Heart Foundation - University College London |
| See Also | http://wiki.geneontology.org/index.php/BHF-UCL |
| URI |
http://nextprot.org/rdf/source/Bgee
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information imported from dataBase for Gene Expression Evolution |
| URI |
http://nextprot.org/rdf/source/CACAO
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Community Assessment of Community Annotation with Ontologies |
| See Also | http://gowiki.tamu.edu/wiki/index.php/Category:CACAO |
| URI |
http://nextprot.org/rdf/source/CAFA
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Critical Assessment of Protein Function Annotation. |
| See Also | http://biofunctionprediction.org/cafa/ |
| URI |
http://nextprot.org/rdf/source/Cellosaurus
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by neXtProt from the Cellosaurus |
| URI |
http://nextprot.org/rdf/source/ComplexPortal
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | A Manually curated, encyclopaedic resource of macromolecular complexes from a number of key model organisms |
| See Also | https://www.ebi.ac.uk/complexportal |
| URI |
http://nextprot.org/rdf/source/Cosmic
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Catalogue Of Somatic Mutations In Cancer |
| See Also | http://www.sanger.ac.uk/genetics/CGP/cosmic/ |
| URI |
http://nextprot.org/rdf/source/DFLAT
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Developmental FunctionaL Annotation at Tufts |
| See Also | http://dflat.cs.tufts.edu/data.htm |
| URI |
http://nextprot.org/rdf/source/DrugBank
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Drug and drug target database |
| See Also | http://www.drugbank.ca/ |
| URI |
http://nextprot.org/rdf/source/Dyp
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted form the Dyp database |
| URI |
http://nextprot.org/rdf/source/ENYO
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | ENYO Pharma, a clinical stage Biotech with a unique drug discovery engine inspired by viruses |
| See Also | http://www.enyopharma.com/ |
| URI |
http://nextprot.org/rdf/source/Ensembl
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information imported from Ensembl |
| See Also | http://www.ensembl.org |
| URI |
http://nextprot.org/rdf/source/FlyBase
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | A database for Drosophila genetics and molecular biology. |
| See Also | http://flybase.org/ |
| URI |
http://nextprot.org/rdf/source/GDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Human Genome Database |
| See Also | http://www.gdb.org |
| URI |
http://nextprot.org/rdf/source/GFP-cDNAEMBL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | GFP-cDNA Localisation Project (EMBL) |
| URI |
http://nextprot.org/rdf/source/GOC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | inferred annotations from GO OBO v1.2 |
| See Also | http://www.geneontology.org |
| URI |
http://nextprot.org/rdf/source/GO_central
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Reference Genome Annotation Project |
| See Also | http://geneontology.org/page/reference-genome-annotation-project |
| URI |
http://nextprot.org/rdf/source/GlyConnect
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | A platform integrating sources of information to help characterise the molecular components of protein glycosylation |
| See Also | https://glyconnect.expasy.org/ |
| URI |
http://nextprot.org/rdf/source/HGNC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | HUGO Gene Nomenclature Committee |
| See Also | http://www.gene.ucl.ac.uk/nomenclature |
| URI |
http://nextprot.org/rdf/source/HGNC-UCL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | HGNC gene annotation from University College of London |
| See Also | https://www.ucl.ac.uk/cardiovascular/research/pre-clinical-and-fundamental-science/functional-gene-annotation/hgnc-gene-annotation |
| URI |
http://nextprot.org/rdf/source/Human_protein_atlas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Expression data imported from Human protein atlas |
| See Also | http://www.proteinatlas.org |
| URI |
http://nextprot.org/rdf/source/IntAct
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by IntAct |
| URI |
http://nextprot.org/rdf/source/InterPro
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by InterPro |
| URI |
http://nextprot.org/rdf/source/KEGG_PTW
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Link to KEGG pathways |
| URI |
http://nextprot.org/rdf/source/LIFEdb
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | LIFEdb |
| See Also | http://www.lifedb.de |
| URI |
http://nextprot.org/rdf/source/MDATA_0004_2011
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the submission MDATA_0004_2011 |
| URI |
http://nextprot.org/rdf/source/MDATA_0023_2012
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Proteomics information extracted from the submission MDATA_0023_2012 |
| URI |
http://nextprot.org/rdf/source/MDATA_0033_2013
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the submission MDATA_0033_2013 |
| URI |
http://nextprot.org/rdf/source/MGI
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Data from the Mouse orthologs database |
| URI |
http://nextprot.org/rdf/source/MTBbase
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Collection and Refinement of Physiological Data on Mycobacterium tuberculosis |
| See Also | http://www.ark.in-berlin.de/Site/MTBbase.html |
| URI |
http://nextprot.org/rdf/source/MassIVE
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Human peptide sequences and identifiers from MassIVE |
| See Also | https://massive.ucsd.edu/ |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Adrenal_Gland
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Adrenal Gland |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Blood_Cells
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Blood Cells |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Blood_Plasma
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Blood Plasma |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Blood_Vessel
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Blood Vessel |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Bone
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Bone |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Brain
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Brain |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Breast
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Breast |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Blood_Cells
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Blood Cells |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Bone
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Bone |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Brain
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer, Brain |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Breast
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer, Breast |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Blood
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Blood |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Bone
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines Bone |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Brain
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Brain |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Breast
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Breast |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Colon
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Colon |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Digestive_System
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines Digestive System |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Liver
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Liver |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Lung
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Lung |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Other_Anatomical_Entities
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Other Anatomical Entities |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Other_Female_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Other Female Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Other_Gestational_Structure
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Other Gestational Structure |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Ovary
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines, Ovary |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Cell_Lines_Skin
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Cell Lines Skin |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Digestive_System
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Digestive System |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Kidney
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer, Kidney |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Liver
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer, Liver |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Lung
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer, Lung |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Other_Male_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer, Other Male Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Ovary
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer, Ovary |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Skin
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer Skin |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cancer_Urinary_Bladder
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cancer, Urinary Bladder |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Cerebrospinal_Fluid
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Cerebrospinal Fluid |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Digestive_System
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Digestive System |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Eye
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Eye |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Heart
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Heart |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Kidney
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Kidney |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Liver
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Liver |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Lung
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Lung |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Muscle
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Muscle |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Non_Cancer_Cell_Lines
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Non Cancer Cell Lines |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Olfactory_System
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Olfactory System |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Oocyte
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Oocyte |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Other_Anatomical_Entities
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Other Anatomical Entities |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Other_Female_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Other Female Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Other_Gestational_Structure
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Other Gestational Structure |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Other_Male_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Other Male Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Other_Respiratory_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Other Respiratory Organ |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Ovary
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Ovary |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Pancreas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Pancreas |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Pituitary_Gland
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Pituitary Gland |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Placenta
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Placenta |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Skin
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Skin |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Sperm
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Sperm |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Spleen
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Spleen |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Testis
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Testis |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Thyroid_Gland
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Thyroid Gland |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Ureter
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Ureter |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Urinary_Bladder
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Urinary Bladder |
| URI |
http://nextprot.org/rdf/source/MassIVE_human_Urine
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by MassIVE, project Urine |
| URI |
http://nextprot.org/rdf/source/NTNU_SB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | NTNU_SB ontology |
| URI |
http://nextprot.org/rdf/source/NextProt
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated automacally or manually by neXtProt |
| URI |
http://nextprot.org/rdf/source/Orphanet
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Orphanet; a database dedicated to information on rare diseases and orphan drugs |
| See Also | http://www.orpha.net/consor/cgi-bin/home.php?Lng=GB |
| URI |
http://nextprot.org/rdf/source/PINC
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Proteome Inc. |
| URI |
http://nextprot.org/rdf/source/PMID_20140087
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 20140087 |
| URI |
http://nextprot.org/rdf/source/PMID_20570859
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 20570859 |
| URI |
http://nextprot.org/rdf/source/PMID_20687582
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 20687582 |
| URI |
http://nextprot.org/rdf/source/PMID_20797634
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 20797634 |
| URI |
http://nextprot.org/rdf/source/PMID_20972266
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 20972266 |
| URI |
http://nextprot.org/rdf/source/PMID_21139048
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 21139048 |
| URI |
http://nextprot.org/rdf/source/PMID_21645671
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 21645671 |
| URI |
http://nextprot.org/rdf/source/PMID_21890473
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 21890473 |
| URI |
http://nextprot.org/rdf/source/PMID_22148984
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 22148984 |
| URI |
http://nextprot.org/rdf/source/PMID_22865923
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 22865923 |
| URI |
http://nextprot.org/rdf/source/PMID_23236377
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 23236377 |
| URI |
http://nextprot.org/rdf/source/PMID_23266961
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 23266961 |
| URI |
http://nextprot.org/rdf/source/PMID_23584533
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 23584533 |
| URI |
http://nextprot.org/rdf/source/PMID_23955771
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 23955771 |
| URI |
http://nextprot.org/rdf/source/PMID_24129315
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 24129315 |
| URI |
http://nextprot.org/rdf/source/PMID_25038526
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 25038526 |
| URI |
http://nextprot.org/rdf/source/PMID_25218447
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information extracted from the PubMed publication 25218447 |
| URI |
http://nextprot.org/rdf/source/ParkinsonsUK-UCL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Parkinsons Disease Gene Ontology Initiative |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Human peptide sequences and identifiers from PeptideAtlas |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Adrenal_Gland
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Adrenal Gland |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Blood_Cells
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Blood Cells |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Blood_Plasma
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Blood Plasma |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Brain
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Brain |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Breast
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Breast |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Adrenal_Gland
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Adrenal Gland |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Brain
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Brain |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Breast
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Breast |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Blood
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Blood |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Bone
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Bone |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Brain
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Brain |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Breast
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Breast |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Digestive_System
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Digestive System |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Kidney
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Kidney |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Liver
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Liver |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Lung
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Lung |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Other_Anatomical_Entities
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Other Anatomical Entities |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Other_Female_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Other Female Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Other_Male_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Other Male Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Ovary
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Ovary |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Pancreas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Pancreas |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Skin
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Skin |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Cell_Lines_Urinary_Bladder
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer Cell Lines, Urinary Bladder |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Digestive_System
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Digestive System |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Kidney
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Kidney |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Liver
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Liver |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Lung
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Lung |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Other_Anatomical_Entities
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Other Anatomical Entities |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Other_Female_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Other Female Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Other_Male_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Other Male Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Ovary
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Ovary |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Pancreas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Pancreas |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Skin
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Skin |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_Urinary_Bladder
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer, Urinary Bladder |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cancer_and_Non_Cancer_Cell_Lines
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cancer and Non Cancer Cell Lines |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Cerebrospinal_Fluid
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Cerebrospinal Fluid |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Digestive_System
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Digestive System |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Eye
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Eye |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Heart
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Heart |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Kidney
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Kidney |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Liver
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Liver |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Lung
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Lung |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Muscle
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Muscle |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Non_Cancer_Cell_Lines
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Non Cancer Cell Lines |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Olfactory_System
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Olfactory System |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Oocyte
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Oocyte |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Other_Anatomical_Entities
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Other Anatomical Entities |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Other_Female_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Other Female Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Other_Gestational_Structure
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Other Gestational Structure |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Other_Male_Reproductive_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Other Male Reproductive Organ |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Other_Respiratory_Organ
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Other Respiratory Organ |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Ovary
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Ovary |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Pancreas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Pancreas |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Pituitary_Gland
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Pituitary Gland |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Placenta
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Placenta |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Skin
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Skin |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Sperm
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Sperm |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Spleen
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Spleen |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Testis
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Testis |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Thyroid_Gland
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Thyroid Gland |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Ureter
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Ureter |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Urinary_Bladder
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Urinary Bladder |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_Urine
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project Urine |
| URI |
http://nextprot.org/rdf/source/PeptideAtlas_human_phosphoproteome
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information generated by PeptideAtlas, project human phosphoproteome |
| URI |
http://nextprot.org/rdf/source/RHEA
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | RHEA |
| See Also | https://www.rhea-db.org/ |
| URI |
http://nextprot.org/rdf/source/Reactome
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Reactome - a knowledgebase of biological pathways and processes |
| See Also | http://www.reactome.org/ |
| URI |
http://nextprot.org/rdf/source/SGD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Saccharomyces Genome Database |
| See Also | http://www.yeastgenome.org/ |
| URI |
http://nextprot.org/rdf/source/SRMAtlas
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Human peptide sequences and identifiers from SRMAtlas |
| URI |
http://nextprot.org/rdf/source/SYSCILIA_CCNET
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | A systems biology approach to dissect cilia function. Imported from GOA. |
| See Also | http://syscilia.org/ |
| URI |
http://nextprot.org/rdf/source/SynGO
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Synapse Gene Ontology and Annotation Initiative. |
| See Also | http://www.geneontology.org/page/syngo-synapse-biology |
| URI |
http://nextprot.org/rdf/source/SynGO-UCL
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | The Synapse Gene Ontology and Annotation Initiative at UCL. |
| See Also | http://www.geneontology.org/page/syngo-synapse-biology |
| URI |
http://nextprot.org/rdf/source/TCDB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Transport Classification Database |
| See Also | http://www.tcdb.org/ |
| URI |
http://nextprot.org/rdf/source/Uniprot
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Information imported from Uniprot |
| See Also | http://www.uniprot.org |
| URI |
http://nextprot.org/rdf/source/WB
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | A comprehensive resource for genetics, genomics and biology of C. elegans and related nematodes |
| See Also | https://www.wormbase.org |
| URI |
http://nextprot.org/rdf/source/YuBioLab
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Center for Bioinformatics and Genomics Systems Engineering (CBGSE), Department of Electrical and Computer Engineering, Texas A&M University |
| See Also | https://yubiolab.wordpress.com/ |
| URI |
http://nextprot.org/rdf/source/dbSNP
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Short genetic variations database |
| See Also | file:///Users/pmichel/Documents/workspace-mars/nextprot-scripts/src/polish-rdf/www.ncbi.nlm.nih.gov/SNP/ |
| URI |
http://nextprot.org/rdf/source/dictyBase
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Dictyostelium discoideum |
| See Also | http://dictybase.org/ |
| URI |
http://nextprot.org/rdf/source/gnomAD
|
|---|---|
| Is Defined By | http://nextprot.org/rdf |
| Description | Genome aggregation database |
| See Also | https://gnomad.broadinstitute.org/ |
http://nextprot.org/rdfhttp://nextprot.org/rdf#https://brickschema.org/schema/Brick#http://www.w3.org/ns/csvw#http://nextprot.org/rdf/db/http://purl.org/dc/elements/1.1/http://purl.org/dc/dcam/http://www.w3.org/ns/dcat#http://purl.org/dc/dcmitype/http://purl.org/dc/terms/http://usefulinc.com/ns/doap#http://xmlns.com/foaf/0.1/http://www.w3.org/ns/odrl/2/http://www.w3.org/ns/org#http://www.w3.org/2002/07/owl#http://www.w3.org/ns/dx/prof/http://www.w3.org/ns/prov#http://purl.org/linked-data/cube#http://www.w3.org/1999/02/22-rdf-syntax-ns#http://www.w3.org/2000/01/rdf-schema#https://schema.org/http://www.w3.org/ns/shacl#http://www.w3.org/2004/02/skos/core#http://www.w3.org/ns/sosa/http://www.w3.org/ns/ssn/http://www.w3.org/2006/time#http://purl.uniprot.org/core/http://purl.org/vocab/vann/http://rdfs.org/ns/void#http://www.w3.org/2001/XMLSchema#| c | Classes |
| op | Object Properties |
| fp | Functional Properties |
| dp | Data Properties |
| ap | Annotation Properties |
| p | Properties |
| ni | Named Individuals |